Fig. 5.
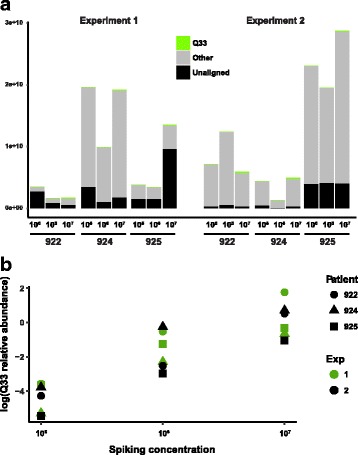
Spiking of faecal samples with an exogenous phage enables absolute quantification of faecal viruses by metagenomic sequencing. a Percentage of reads aligning to the spiked lactococcal bacteriophage Q33 genome depends linearly on the spiked phage titre in faecal sample. b Spiking of faecal samples at concentration of 106 pfu g− 1 seems to be optimal in terms of percentage of reads aligning to the internal standard phage genome
