FIG 5.
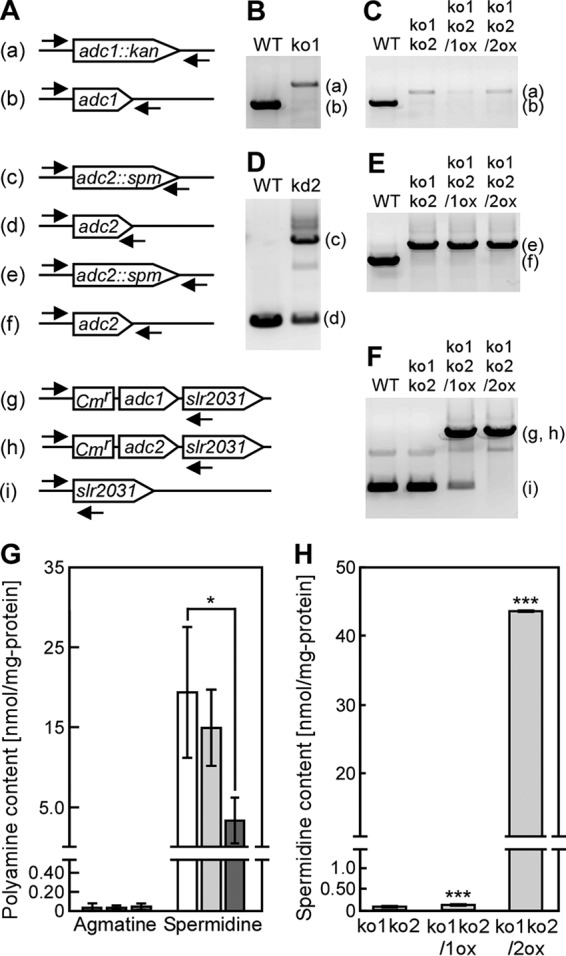
Spermidine contents in Synechocystis mutants. (A to F) The arrows indicate the positions and orientation of the primers for PCR to confirm the successful disruption or reintroduction of adc1 and adc2 in the Synechocystis genome. The letters on the left correspond to the PCR bands in panels B to F. The overexpression cassette of adc1 (g) or adc2 (h) was integrated at a neutral site, the slr2031 gene. Disruption of the genes in the insertion mutants ko1, kd2, and ko1ko2, obtained by repeated homogeneity segregation, was determined by the presence of a PCR product of the expected length. Note that for adc2 no complete disruption could be obtained (see the text for details). (G) Agmatine and spermidine content of the wild-type (WT) (white bars), ko1 (light gray bars), and ko2 (dark gray bars) strains were measured in cells grown for 5 days in medium without added NaCl. The spermidine content data for the wild type are the same as those shown in Fig. 2. Each value corresponds to the mean ± SD (n = 4). Significant differences between the WT and each mutant were analyzed by Student's t test (*, P < 0.05). No significant differences (P > 0.05) were found in agmatine content by Student's t test. (H) The spermidine contents of ko1ko2, ko1ko2/1ox, and ko1ko2/2ox strains were measured in cells grown for 7 days in medium without added NaCl. Each value corresponds to the mean ± SD (n = 3). Significant differences between ko1ko2 and ko1ko2/1ox or ko1ko2/2ox were analyzed by Student's t test (***, P < 0.001).
