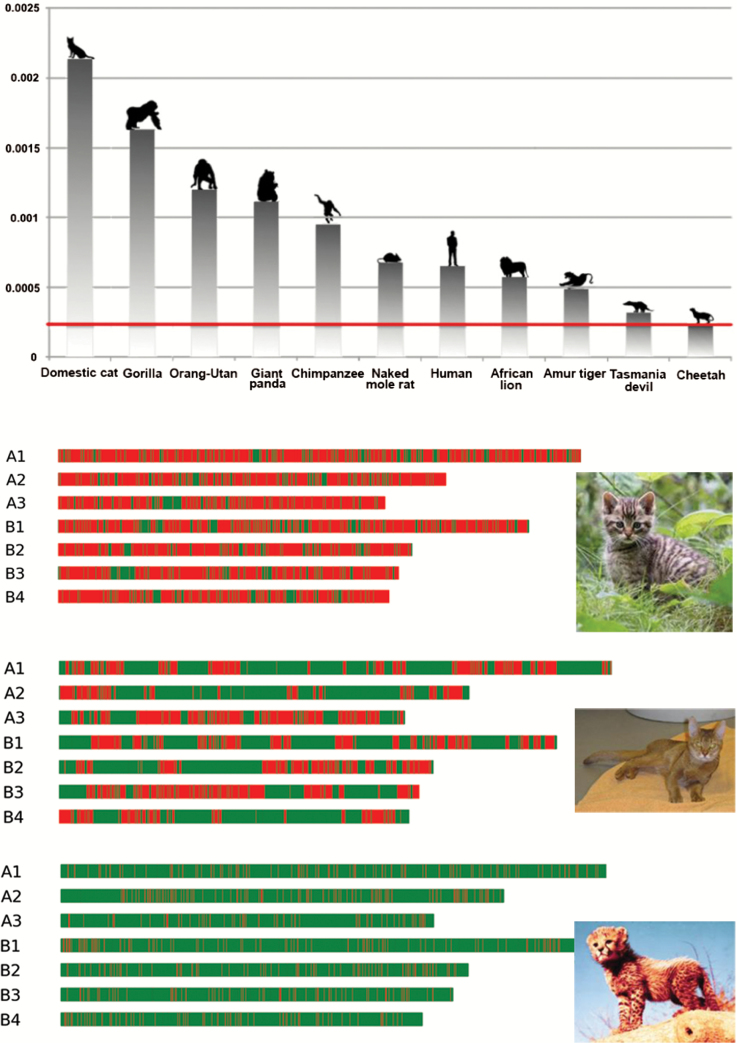
Figure 1.
(A) Estimates of diversity in the cheetah genome relative to other mammal genomes. The rate of single nucleotide variation (SNV) (x axis) for each individual was estimated using all variant positions, and repetitive regions were not filtered. (B) The genome of Boris, an outbred feral domestic cat living in St. Petersburg, Russia (top) is compared to Cinnamon, a highly inbred Abyssinian cat [Fca-6.2 reference for domestic cat genome sequence (Tamazian et al. 2014)] (middle) and Chewbacca, a captive cheetah (bottom) (Dobrynin et al. 2015). The first 7 chromosome homologues of the genomes of Boris, Cinnamon, and Chewbacca are displayed for direct comparison. Approximately 15000 regions of 100 Mb across the genome for each species were assessed for SNVs. Regions of high variability (>40 SNVs/100 kbp) are colored red (dark gray); highly homozygous regions (≤40 SNVs/100 kbp) are colored green (light gray). The cheetah genome is composed of 93% homozygous stretches. Reprinted from Dobrynin et al. (2015) with permission.