Abstract
Mating induces a multitude of changes in female behavior, physiology, and gene expression. Interactions between female and male genotype lead to variation in post-mating phenotypes and reproductive success. So far, few female molecules responsible for these interactions have been identified. Here, we used Drosophila melanogaster from 5 geographically dispersed populations to investigate such female × male genotypic interactions at the female transcriptomic and phenotypic levels. Females from each line were singly-mated to males from the same 5 lines, for a total of 25 combinations. Reproductive output and refractoriness to re-mating were assayed in females from the 25 mating combinations. Female × male genotypic interactions resulted in significant differences in these post-mating phenotypes. To assess whether female × male genotypic interactions affect the female post-mating transcriptome, next-generation RNA sequencing was performed on virgin and mated females at 5 to 6 h post-mating. Seventy-seven genes showed strong variation in mating-induced expression changes in a female × male genotype-dependent manner. These genes were enriched for immune response and odorant-binding functions, and for expression exclusively in the head. Strikingly, variation in post-mating transcript levels of a gene encoding a spermathecal endopeptidase was correlated with short-term egg production. The transcriptional variation found in specific functional classes of genes might be a read-out of female × male compatibility at a molecular level. Understanding the roles these genes play in the female post-mating response will be crucial to better understand the evolution of post-mating responses and related conflicts between the sexes.
Keywords: female × male genotype interactions, Global Diversity Lines, immune system, reproduction, sexual conflict, transcriptome
In sexually reproducing organisms, reproduction is the result of complex interactions between females and males at the organismal, cellular, and molecular levels. In addition, reproductive success varies depending on the genotypes of the female and her mate. Female and male genotypic effects are often nonadditive in their impact on mating. Genotypic interactions between females and males can affect pre- and post-copulatory traits that in turn influence reproductive success. For example, interactions between female and male genotype were found to affect female mating rate, fecundity, refractoriness to re-mating (i.e., the likelihood that a previously mated female will re-mate) and sperm competition outcome (Clark and Begun 1998; Clark et al. 1999; Andrés and Arnqvist 2001; Nilsson et al. 2003; Lawniczak and Begun 2005; Chow et al. 2010, 2013; Giardina et al. 2011; Reinhart et al. 2015). Furthermore, female × male genotypic interactions can mediate gametic incompatibility between species (Phadnis and Orr 2009; Satyaki et al. 2014; Tang and Presgraves 2015). Allelic variation in genes important for reproduction largely underlies these female × male genotypic interactions.
Often genes involved in reproductive processes show accelerated rates of evolution; this is thought to be triggered by pressures arising from sexual selection and sexual conflict (Swanson et al. 2001, 2004; Swanson and Vacquier 2002; Panhuis and Swanson 2006). Post-copulatory sexual selection potentially mediates co-evolution between females and males from the same population, and this selective force acts to optimize reproductive success. On the other hand, reproductive genes can also be impacted by sexual conflict, as female and male reproductive interests do not always align (Birkhead and Pizzari 2002). For example, female refractoriness to re-mating is beneficial for the first male to mate. However, it is not necessarily advantageous for the female, as females might benefit from mating with and acquiring sperm from different males. Sexually antagonistic selection can prompt an arms race between females and males, leading each sex to move towards their own reproductive optimum (Sirot et al. 2015). Female × male co-evolution within populations can promote inter-population divergence of molecules required for reproduction. Divergence of reproductive molecules is hypothesized to lead to “miscommunication” between females and males from isolated populations, eventually resulting in reduced reproductive output and the generation of reproductive barriers that may ultimately lead to speciation (Panhuis et al. 2001; Kirkpatrick and Ravigné 2002; Ritchie 2007).
In D. melanogaster, male-derived molecules have been identified that govern female × male genotypic interactions that affect reproductive phenotypes. After mating, females undergo behavioral, physiological, morphological and gene expression changes, that are collectively termed “post-mating responses” (Lawniczak and Begun 2004; McGraw et al. 2004, 2008; Mack et al. 2006; Kapelnikov et al. 2008; Avila et al. 2011; Apger-McGlaughon and Wolfner 2013; Heifetz et al. 2014; Mattei et al. 2015; Reiff et al. 2015). Female post-mating responses are mediated in part by male-derived seminal fluid proteins (Avila et al. 2011). For example, females mated to transgenic males that lack specific seminal fluid proteins, show differences in post-mating transcript abundances, as compared to females mated to wildtype males with a full complement of seminal fluid proteins (McGraw et al. 2004, 2008; Domanitskaya et al. 2007; Gioti et al. 2012). Additionally, polymorphisms in genes encoding seminal fluid proteins impact female post-mating responses and the male’s reproductive success (Clark et al. 1995; Prout and Clark 1996; Hughes 1997; Clark et al. 2000; Fiumera et al. 2005, 2006; Greenspan and Clark 2011; Lüpold et al. 2012; Zhang et al. 2013). Thus, male seminal fluid proteins represent a major molecular component in the reproductive interactions that affect post-mating phenotypes in D. melanogaster.
With the exception of the female receptor for the seminal fluid protein Sex Peptide (SP) (Yapici et al. 2008), the female proteins that are involved in these interactions remain poorly understood. One study aimed to address this gap by quantifying female transcriptional responses after mating with a male from the same isogenic strain versus a male from a different strain (McGraw et al. 2009). No female transcripts responded significantly differently to mating depending on male genotype, however there was limited divergence between the 2 strains that were used (McGraw et al. 2009). Still, identifying female genes involved in female × male interactions is essential to understanding the molecular and physiological mechanisms behind variation in post-mating responses. Furthermore, establishing the female genetic basis that underlies female × male interactions is necessary to shed light on the biological processes that play a role in the evolution of post-mating responses. Particularly interesting are those that are affected by sexual conflict, as they potentially advance reproductive isolation and speciation.
In this study, we aimed to identify female genes involved in female × male genotypic interactions, by measuring post-mating transcriptional changes in females mated to males from diverged populations. We exploited natural genetic variation by using 5 lines drawn from the Global Diversity Lines, a panel of 84 D. melanogaster inbred lines collected from 5 geographically dispersed populations (Beijing, Ithaca, Netherlands, Tasmania, and Zimbabwe) (Grenier et al. 2015). Using females and males from 5 Global Diversity Lines, we used a 5 × 5 mating scheme to produce 25 different mating combinations. We measured post-mating gene expression changes in whole females using RNAseq and evaluated the effects of female genotype, male genotype, and female × male genotypic interactions on post-mating transcriptional variation. To assess whether variation in post-mating transcription affected reproductive success, we also measured physiological post-mating responses (fecundity and hatchability) and a behavioral post-mating response (female refractoriness to re-mating) for the 25 mating combinations.
We found evidence for extensive variation due to female × male genotypic interactions in all post-mating responses that we investigated. In particular, female × male genotypic interactions influenced classes of genes that might be predictive of female × male compatibility at the molecular level.
Materials and Methods
Lines of Drosophila melanogaster and Husbandry
Five D. melanogaster inbred lines were used. These lines are derived from 5 geographically dispersed populations (Global Diversity Lines Beijing 04; Ithaca 16; Netherlands 01; Tasmania 01; and Zimbabwe 184—the latter line was collected in Africa, but turned out to be a recent migrant) (Grenier et al. 2015). These 5 lines were chosen because of their low levels of heterozygosity, which should limit within-line phenotypic and transcriptional variation. Flies were maintained on standard yeast/glucose media on a 12 h light/dark cycle at 25 °C. Virgin females and males were aged 3–5 days in single-sex groups before the start of each experiment. For fecundity and hatchability assays, females were supplemented with live yeast during aging and for the duration of the assays.
Mating Scheme and Sample Collection
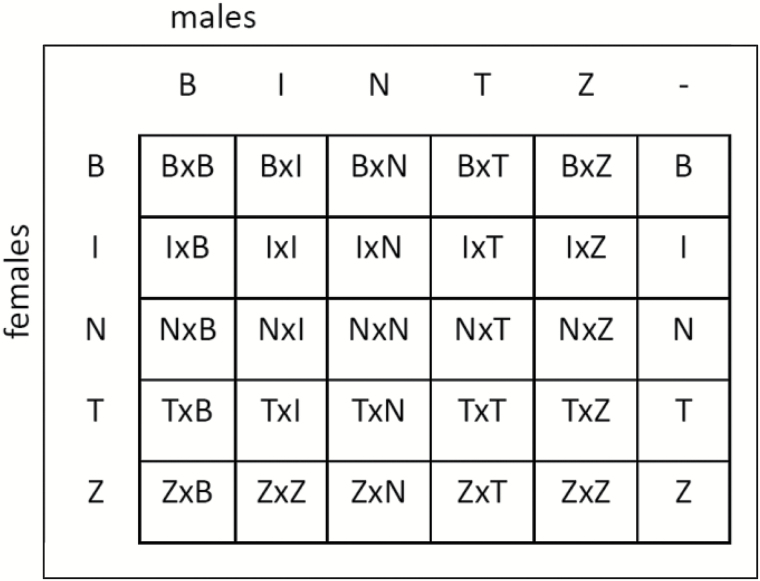
Virgin females from each line were singly-mated to virgin males from each of the 5 inbred lines, similar to a 5 × 5 full factorial design (Figure 1). We refer to each cross by the female used, followed by the male used, and replicate number. All matings were observed and males were removed at the end of copulation. For RNAseq and qRT-PCR, mated females were flash frozen 5 to 6 h after the start of mating. Age-matched virgin females were flash-frozen in parallel. This timepoint was chosen to ensure a robust response with transcriptional changes of larger magnitude, compared to earlier time points (Mack et al. 2006; McGraw et al. 2008). For RNAseq, 3 independent biological replicates were generated for each of the 25 mating combinations and for virgin females of each genotype (90 samples total). Flies from each replicate were collected from separate bottles, and matings for all 3 replicates were set up simultaneously. RNA was extracted from 5 to 10 pooled females per replicate. Note that this experimental design resembles a diallel cross. However, gene expression was measured in the females after mating, rather than in the F1 progeny.
Figure 1.
Crossing scheme for the 5 Drosophila melanogaster inbred lines (B = Beijing, I = Ithaca, N = Netherlands, T = Tasmania, Z = Zimbabwe). The last column represents virgin females. Cross names list the female’s genotype first. After the mating within each cell of the table, RNA was isolated from females only.
For qRT-PCR, 3 to 4 independent biological replicates were collected, with 10 females pooled per replicate. Three of the genes tested using qRT-PCR are involved in the immune response (Dro, Def, AttB). We were interested in determining if the expression of these immune genes was affected by female and male genotype. Because immune gene expression is also highly dependent on unmeasured environmental factors such as wild microbial contamination (Gibson 2008), independent biological replicates were collected from 2 independent cultures of flies of the same genotype, raised in parallel (Supplementary Figure S12).
Transcript Detection
RNA was extracted from whole flies using Trizol (Rio et al. 2010). Whole flies were used because we did not have prior expectations of which tissue(s) might be most important, and because previous studies had shown that even spermathecae-specific genes were readily detected in whole-fly transcriptome analyses (e.g., McGraw et al. 2008). RNAseq libraries were prepared using Illumina’s Truseq RNA Library Preparation Kit v2 (cat# RS-122–2001, RS-122–2002). Samples were sequenced in a single-end 100 bp run on a HiSeq2000, at the Genomics Facility in the Cornell Biotechnology Resource Center. For qRT-PCR, RNA was DNase-treated using RQ1 RNase-Free DNase from Promega and cDNA was synthesized using Clontech SMARTScribeTM Reverse Transcriptase. Quantitative PCR was done using the LightCycler 480 SYBR Green I Master from Roche and a Roche LightCycler 480 instrument. Rp49 was used as a control gene for normalization in qRT-PCR assays (Ponton et al. 2011). Rp49 transcript levels were found not to change after mating in our dataset (Supplementary Table S1). Primer sequences were designed using NCBI primer-BLAST (Supplementary Table S2). Results were analyzed using the ΔΔCt method (Livak and Schmittgen 2001), based on 3 technical replicates for each biological replicate.
RNAseq Data Processing and Analysis
Read Processing and Alignment
FastQC was used to assess the quality of the libraries (http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc). One library (I female × Z male, replicate-2) was discarded because it contained a very low number of reads (106 087 reads compared to an average of 21 million reads per library). Bases at the 5′ end of reads with a phred score lower than 20 were removed using Trimmomatic (Bolger et al. 2014). Reads shorter than 32 bp were discarded. Reads were aligned to the D. melanogaster reference genome (dm3) using TopHat2 (Kim et al. 2013). We used the default settings and did not include novel splice discovery, similar to Trapnell et al. (2012). HTseq-count (Anders et al. 2015) was used to determine the raw number of read counts per gene.
Sample Quality Control
Biological replicates were compared using MA plots and MDS plots, which indicated 10 outlier replicates (B × I-2; B × N-2; B × Z-1; I × I-3; I × N-1; I × T-3; I × Z-1; I × Z-3; N × B-2; N × I-2; Supplementary Figures S1–S5). Eight of the 10 outlier replicates did not cluster as expected by female genotype or mating status. Two of the 10 outlier replicates showed 2-fold or higher differences in expression for over 1000 genes, relative to their biological replicates. These 10 replicates were removed from the dataset before filtering out lowly expressed genes. Because all 3 I × Z samples were removed due to low quality or as outliers, the I × Z combination was completely removed from our dataset. This left 24 mating combinations whose gene expression was to be analyzed.
Differential Expression Analyses
EdgeR was used to analyze differential mRNA abundance (McCarthy et al. 2012; Robinson et al. 2010). Read counts were normalized using the CPM (counts per million) function with TMM normalization, to control for size differences among libraries. Based on the normalized counts, lowly expressed genes were removed: a gene was kept in the dataset if it had a CMP > 3 in at least 3 samples. This filtered the dataset down from 14 522 genes to 9484 genes (Supplementary Figure S6).
Four distinct differential expression analyses were conducted, using linear models, each with its own design matrix. All models were controlled for batch effects, because MDS plots demonstrated a clustering of samples that were processed simultaneously (Supplementary Figure S4). For each linear model, contrasts were set up to find differentially expressed genes for the comparisons of interest. Differential expression analyses were performed to answer 4 distinct questions (Supplementary Figure S7):
-
Which genes respond to mating regardless of female or male genotype?
All mated females were compared with all virgin females. (1 contrast total)
-
Which genes respond to mating in a female × male genotype interaction-dependent manner?
The response to mating in a female mated to a particular male was compared with the average response to mating across all combinations of females and males. (24 contrasts total; we did not include the I × Z combination)
-
Which genes respond to mating in a female genotype-dependent manner?
The response to mating in a particular female genotype was compared with the average response to mating across all females. (5 contrasts total)
-
Which genes respond to mating in a male genotype-dependent manner?
The response to mating in females mated to a particular male genotype was compared to the average response to mating across all females mated to all males. (5 contrasts total)
For questions 2, 3, and 4, it is important to note that we were not interested in directly comparing gene expression between females from different lines. Instead, we were interested in detecting differences in the response to mating. Because of this, we always compared females with their respective virgins, before comparing between lines. For each of the 35 contrasts, we retrieved genes with q-values < 0.05 (P-values corrected for multiple testing using the Benjamini-Hochberg method; Benjamini and Hochberg 1995; raw P-value quantile-quantile plots: Supplementary Figures S8–S11). Flybase and FlyAtlas were used to retrieve information on gene function and tissue-specific expression (Chintapalli et al. 2007; Attrill et al. 2016). DAVID was used to test for enrichment of functional classes among the differentially expressed genes (Huang et al. 2008, 2009). A 5 by 5 factorial ANOVA was used as a different method to address the roles of female and male genotype on post-mating gene expression changes (Supplementary Information p. 15).
Permutation Tests
Permutation tests were performed to ensure that the number of differentially regulated genes detected for questions 2, 3, and 4 differed significantly from the number of differentially regulated genes found by chance. Random sampling was done in R to permute the RNAseq dataset 500 times. Permutations were done within replicate 1, 2, or 3 to still permit for batch effect control in the linear models. The edgeR analyses for questions 2, 3, and 4 were repeated 500 times. For each of the 34 contrasts, we calculated the likelihood of finding a number of differentially regulated genes equal to or larger than the number of differentially regulated genes observed for that contrast based on the original dataset.
Wolbachia
Four of the 5 lines we used carry the bacterial endosymbiont Wolbachia pipientis. Only the line from the Netherlands is uninfected. Additional analyses were performed to assess whether the female × male genotypic interactions we observed were due to the presence or absence of Wolbachia. The results and discussion of these analyses can be found in Supplementary Information, p. 16–18.
Phenotypic Assays
Fecundity and Hatchability Assays
Singly-mated females were allowed to lay eggs for 24 h and were then transferred to a new vial. This was repeated for a total of 5 days (5 vials per female), and eggs were counted daily as a measure of fecundity. Per-vial hatchability was determined as the proportion of eggs that developed into pupae. A total of 3 independent assays were set up. Egg count and hatchability data were collected from 543 females, yielding an average of 21.7 females for each of the 25 mating combinations. Data from females that died during the experiment, and data from 6 females that produced fewer than 10 eggs over the course of 5 days were excluded. Egg count data were analyzed in R version 3.3.2 using the lme4 and lsmeans packages (Magezi 2015; Lenth 2016). We tested whether the number of eggs produced by a female differed depending on 1) female genotype, 2) male genotype, 3) time, or 4) all possible interactions between these 3 main factors. Data were fitted using a linear mixed effects model, which assumes a normal error distribution (Supplementary Figure S13). To control for repeated measures on the same female (daily egg counts) “female_ID” was included as a random effect. When analyzing the 3 assays separately, comparable results were found. Because of this, all 3 assays were analyzed simultaneously, and “block” was added as an additional random effect to the model. The proportion of hatched eggs was analyzed using a similar model (Supplementary Figure S14). In these models, i represents the effect of the ith female genotype, j represents the effect of the jth male genotype, k represents the effect of the kth day, l represents the effect of the lth block and m represents the effect of the mth individual female.
Phenotypeijklm ~ femalei + malej + dayk + (femalei * dayk) + (malej * dayk) + (femalei * malej) + (femalei * malej * dayk) + (1|block)l + (1|female_ID)m + ɛijklm
Female Refractoriness to Re-mating
At 24 h and at 4 days after the first mating with a male from a Global Diversity Line, a single 3- to 5-day-old virgin Canton-S male was aspirated into a vial with one mated female. Pairs of females and males were observed for 1 h, and the number of females that started mating with the Canton-S male within that hour was recorded. Five assays were conducted to test refractoriness to re-mating on day 1 after the initial mating, and 4 assays were performed to test refractoriness to re-mating on day 4 after the initial mating. In total, an average of 32 females was tested per female × male combination. We tested whether the number of females that re-mated within 1h differed depending on 1) female genotype, 2) male genotype, or 3) the interaction between female and male genotype. Refractoriness on day 1 and day 4 after mating was analyzed separately. The assays were analyzed using a linear mixed effects model assuming a normal error distribution (Supplementary Figure S15). In these models, i represents the effect of the ith female genotype, j represents the effect of the jth male genotype, and k represents the effect of the kth block.
Proportion re-matedijk ~ femalei + malej + (femalei * malej) + (1|block)k + (1|blockk * femalei) + (1|blockk * malej) + ɛijk
Correlations Between Reproductive Phenotypes and the Transcriptional Response to Mating
Correlations between the transcriptional response to mating and reproductive phenotypes were investigated using a Spearman rank correlation test. As a measure of the transcriptional response to mating, edgeR’s estimates of the log2 fold changes of mated versus virgin females were used for each of the 24 female × male combinations (I × Z was excluded). Correlations were investigated between fold changes and 1) the total number of eggs produced over a period of 5 days, 2) the average number of eggs produced per day, 3) the total number of eggs produced on day 1 after mating, and 4) the proportion of females that re-mated 4 days after the first mating. Correlation tests were performed first with the genes that were found to be differentially regulated depending on an interaction between female and male genotype. Second, correlation tests were done with all 9484 genes in our filtered dataset. Because correlations were examined for each gene independently, P-values were corrected using the Benjamini-Hochberg method (Benjamini and Hochberg 1995).
Results
Transcript Levels of 272 Genes Change Post-Mating Across all Mating Combinations
To identify female × male genotypic interactions that influence post-mating transcriptional changes, gene expression was analyzed in mated females from 24 different mating combinations. Specifically, gene expression was measured in females from 5 diverged lines that were singly-mated to males from these 5 lines (one combination, I × Z, was excluded from our analysis; see Materials and Methods). First, we investigated the overall transcriptional response to mating, averaged across all 24 combinations. We detected 272 differentially expressed genes in mated females universally, regardless of female and male genotypes (Supplementary File 2). Of these 272 genes, 50 were down-regulated and 222 were up-regulated in mated females. Only a minority of these genes underwent a 2-fold or greater change in RNA abundance (7 out of 50 for the down-regulated genes, 25 out of 222 for the up-regulated genes). Gene Ontology (GO) functions of the 50 down-regulated genes include cytoskeleton dynamics, immune response, chitin metabolism, sugar and fatty acid metabolism, and genes with functions in the ovary. Among the 222 up-regulated genes, a large proportion is exclusively or highly expressed in the ovary (64/222 genes). Twenty-four genes are exclusively or predominantly expressed in the digestive system, and 9 are predominantly expressed in the spermathecae (Chintapalli et al. 2007; Attrill et al. 2016). Up-regulated transcripts encode proteins involved in lipid metabolism, odorant binding, protein folding, the endomembrane system, neurogenesis and muscle system processes, the immune response and chitin cuticle structure, consistent with previous studies (Mack et al. 2006; Kapelnikov et al. 2008; McGraw et al. 2008; Dalton et al. 2010).
Transcript Levels of 77 Genes are Differentially Regulated Post-Mating Depending on Interactions Between Female and Male Genotype
Seventy-seven genes responded differently to mating in specific female × male combinations, relative to the average response to mating across all 24 combinations (Figures 2 and 3, Supplementary Figure S16, Supplementary File 3). This was greater than the number of differentially expressed genes found by chance based on permutation tests (Supplementary Table S3). Therefore, the differential expression of these 77 genes likely represents real biological effects caused by female × male genotypic interactions. On average, transcript levels of these 77 genes were 2.3 times more or less abundant in one specific mating combination, relative to the post-mating abundance of those transcripts across all 24 combinations. For the majority of the 77 genes, the female × male genotype interaction was driven by only one mating combination (Figure 2). Only CG8343 (differentially expressed in B × I and B × N) and 10 genes that were differentially expressed in mated Tasmania females (Figure 2) showed differential expression in more than one mating combination.
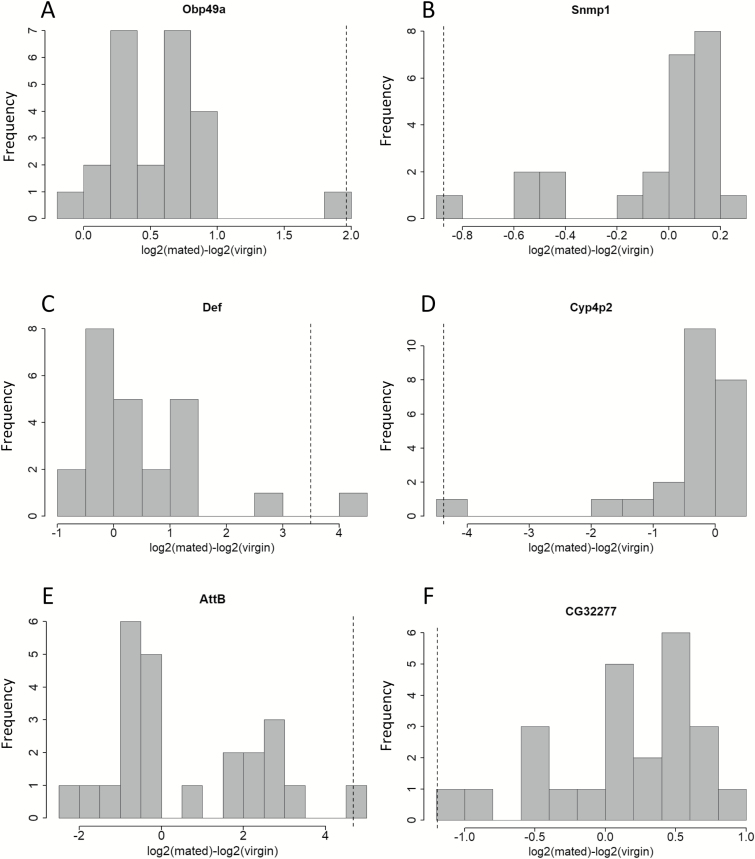
Figure 2.
For 77 genes, the mating-induced transcriptional changes differed depending on interactions between female and male genotype (q < 0.05). Colored cells indicate the 77 genes with respective female × male combinations with significant up- (grey) or down- (black) regulation relative to the average post-mating response across all mating combinations.
Figure 3.
Distribution of post-mating fold changes across all female × male combinations, for 6 genes. The dotted line represents the fold change in the genotype(s) that differed significantly from the average post-mating fold change. A: Obp49a transcript levels were up-regulated higher than average in I × N (q = 0.015). B: Snmp1 transcript levels were down-regulated more than average in T × B (q = 0.048). C: Def transcript levels were up-regulated more than average in T × T (q = 0.19) and T × Z (q = 6 × 10–6). D: Cyp4p2 mRNA levels were down-regulated more in B × N (q = 1.5 × 10–5). E: AttB transcripts were up-regulated more than average in B × I (q = 0.041). F: CG32277 mRNA was down-regulated more than average in B × B (q = 0.006).
Validation of 6 transcripts with post-mating expression changes was tested using qRT-PCR (AttB, Def, Dro, Cyp4p2, CG3088, and Obp49a). These genes were selected for qRT-PCR testing based on their q-value (<0.05) and their fold change after mating (at least 2-fold up or down). QRT-PCR validated the RNAseq results for Def, AttB, Dro, CG3088, and Cyp4p2 (Supplementary Table S6; Supplementary Figures S19A–E and S20), even though Cyp4p2 CPM values were very low (<3) in Beijing females. AttB mRNA levels were higher in B × I relative to the average AttB mRNA levels in the RNAseq dataset, but there was a large disparity in the CPM values of the 2 B × I replicates for AttB (CPM for B × I-1= 487, CPM for B × I-2 = 40). Still, qRT-PCR results confirmed a strong and consistent up-regulation of AttB transcripts in B × I, based on 3 biological replicates. Obp49a mRNA levels increased strongly after mating in I × N based on the RNAseq data, and the 2 biological replicates in the RNAseq dataset were very similar (CPM for I × N-1 = 18, CPM for I × N-3 = 27; Supplementary Table S6, Supplementary Figure S19D). However, only one out of 3 biological replicates for I × N showed a post-mating increase in Obp49a transcript levels in the qRT-PCR experiments.
Female- or Male-Genotype Dependent Changes in Transcript Abundance are Uncommon
In addition to identifying interaction effects, we also assessed whether transcriptional responses to mating differed depending solely on female or male genotype. Only 2 genes were differentially regulated depending on the genotype of the male a female mated with, but these results were not well supported by permutation tests (Supplementary Table S5, Supplementary File 3). Twenty-four genes were differentially regulated in a female genotype-dependent manner, regardless of the male with whom these females mated (Supplementary File 3). One of these genes, Acer, a gene involved in the regulation of sleep (Carhan et al. 2011), was differentially regulated in females from the Ithaca line. However, permutation tests showed that this result for females from the Ithaca line was not different from what could be found by chance (Supplementary Table S4). The remaining 23 genes were differentially regulated in females from the Tasmania line. This number was larger than the number of differentially expressed genes that were found by chance based on permutation tests (Supplementary Table S4). These 23 genes included 2 genes with expression in the ovary (CG12200, CR43837, Chintapalli et al. 2007), 2 genes involved in sensory perception of taste (Ir7a and Gr9a), one gene encoding a spermathecae-specific cytochrome (Cyp12d1-d; Prokupek et al. 2009), and 5 genes with high expression in the digestive system or Malpighian tubules (CG10477, Cyp12a5, CG1139, CG11034, CG17752,Chintapalli et al. 2007). One gene, CG13749, is up-regulated in infected virgin females (Short and Lazzaro 2013), and was significantly down-regulated in our mated females from the Tasmania line. This suggests that some of the virgin samples from the Tasmania line carried a pathogen. Indeed, virgin samples from the Tasmania line (and the Zimbabwe line as well) showed high CPM values for a set of antimicrobial peptides (AMPs) (Supplementary Figure S18). That some of our virgin samples might have carried pathogens suggests that caution might be needed with the interpretation of the post-mating changes observed in females from the Tasmania line.
Specific Tissues and Gene Functional Classes are Affected by Female × Male Interactions at the Transcript Level
The 77 genes that were differentially regulated post-mating depending on female and male genotype tend to fall into specific functional classes, or are highly expressed in particular tissues. These tissues and biological functions likely represent molecular mechanisms that underlie variation in female phenotypic post-mating responses and reproductive success. The 77 genes were significantly enriched in genes encoding proteases (DAVID EASE score 1.1 × 10–3) and immune response genes (DAVID EASE score 1.8 × 10–4). Specifically, 12 of the 77 genes play a role in the immune response (Supplementary Figure S17). These include antimicrobial peptides (AttB, Def, Dro, Drs), 3 endopeptidase inhibitors (Tep1, Tep2, Spn28Dc), 2 proteases (Jon65Aii, CG5909), a peptidoglycan recognition receptor (PGRP-SC2), a protease involved in hemolymph coagulation (CG11313), and a gene with unknown molecular function (edin). Twenty-two of the 77 genes are expressed exclusively or predominantly in the head (Attrill et al. 2016, Chintapalli et al. 2007). These include serine-type endopeptidases (CG7829, CG9676, CG3088), odorant-binding proteins (Opb56g, Obp49a, Obp56h), one gene involved in neurogenesis (CG12158), a G-protein coupled receptor involved in phototransduction (Rh6), an olfactory receptor (Snmp1), and carbohydrate-binding proteins (CG8343, CG11211). The latter 2 are also predicted to function as non-self-recognition proteins in the immune response (Theopold et al. 1999). In addition, 4 of the 77 genes are highly expressed in the ovary, 15 genes have expression bias to the digestive system, 2 genes encoding G-protein coupled receptors have high expression in the thoracic-abdominal ganglion, and 7 genes are highly expressed in the spermathecae (Chintapalli et al. 2007). The latter included the serine-type endopeptidase CG32277, Esp and CG8329. A total of 43 out of the 77 genes were previously reported to respond to mating in Drosophila (McGraw et al. 2004, 2008; Mack et al. 2006; Kapelnikov et al. 2008; Prokupek et al. 2009; Dalton et al. 2010; Bono et al. 2011; Short and Lazzaro 2013; Zhou et al. 2014; Hollis et al. 2016).
Phenotypic Post-Mating Responses are Influenced by Female × Male Genotypic Interactions
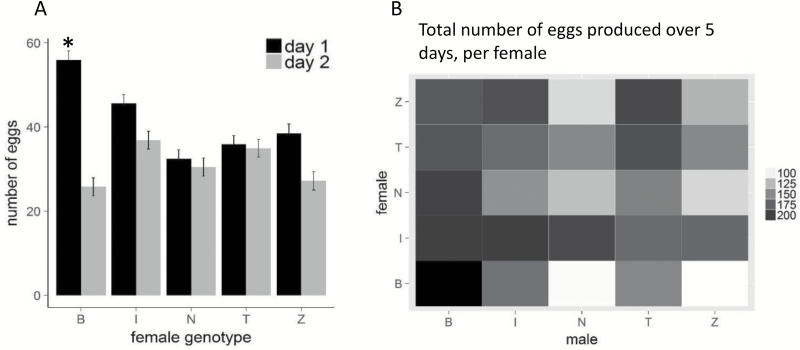
Fecundity, as defined by egg production over the course of 5 days—and the rate at which egg production decreased over time—differed depending on the mating combination (P < 7 × 10–6, Supplementary Figure S21, Supplementary Tables S7 and S8). The strongest interactions occurred with Beijing, Netherlands or Zimbabwe females that mated to males from the Netherlands or Zimbabwe lines. These combinations produced on average 111 (±4) eggs over the course of 5 days, while other combinations produced on average 178 eggs (±4; Figure 4B; Supplementary Figure S21). On day 1 post-mating, strong differences were observed between female genotypes, regardless of male genotype (Supplementary Table S8). On day 1, females from the Beijing line produced on average 56 (± 5) eggs, while other female genotypes produced on average 38 (±5) eggs. This was followed by a rapid decline in egg numbers on day 2 in Beijing females (23 ± 4 eggs), but not in other females (32 ± 4 eggs) (Figure 4A).
Figure 4.
A: Egg production on day 1 and day 2 after mating, for the 5 female genotypes. On day 1 after mating, females from the Beijing line produced significantly more eggs compared to all other females. Due to a rapid decline in egg numbers on day 2, this significant difference disappeared on day 2 after mating (*P < 0.05; error bars indicate standard errors). B: Total number of eggs per female, produced over the course of 5 days, for all 25 mating combinations. Female × male genotype interactions affected the total number of eggs produced over a total of 5 days (P = 1.6 × 10–5; average n for each of the 25 combinations = 21.7).
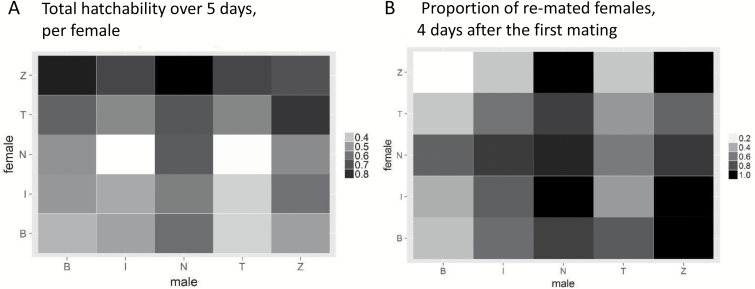
Similar to the fecundity data, the proportion of hatched eggs on a given day, and the decrease in hatchability over time, differed depending on interactions between female and male genotype (P = 0.01; Supplementary Figure S22; Supplementary Tables S9 and S10). Hatchability was consistently high in females from the Zimbabwe line, with an average hatch rate of 77% (±5; Figure 5A). In other females, hatchability varied depending on the genotype of the male, with hatch rates ranging from 38% in B × T, to 79% in T × Z (±5; Figure 5A). These results exclude females from the Netherlands line, the only line not infected with Wolbachia. For females from the Netherlands line who mated to a male that carries Wolbachia, most eggs did not hatch due to unidirectional cytoplasmic incompatibility (Hoffmann et al. 1998). Incomplete incompatibility was observed, whereby the incompatibility was stronger in N × I and N × T crosses. This is consistent with the findings of Poinsot et al. (1998), who showed that the effect of Wolbachia can differ in distinct genetic backgrounds.
Figure 5.
A: Proportion of hatched eggs relative to the total number of eggs produced over the course of 5 days. Female × male genotypic interactions affected hatchability (P = 0.01; average n for each of the 25 combinations = 21.7). B: Proportion of females that re-mated with a standard male, 4 days after the first mating with a male from the Global Diversity Lines. The tendency to re-mate with a standard male on day 4 after the first mating, differed depending on female genotype, and depending on the genotype of the male she mated with for the first mating (P = 2.8 × 10–5; average n for each of the 25 combinations = 32).
Re-mating rates at 24h after the first mating did not differ depending on female genotype, or on the genotype of the first male with whom she mated (Supplementary Table S11). At 4 days after the first mating, female refractoriness to re-mating differed significantly depending on interactions between female and male genotype (P = 0.002, Supplementary Tables S12 and S13). Males from the Beijing line successfully induced refractory behavior in all female genotypes (only 30% of females re-mated, ± 1), except when paired with females from the Netherlands line (71% of females re-mated, ±1; Figure 5B). Males from the Netherlands or Zimbabwe lines were worst at inducing refractory behavior in any female, with a re-mating rate of 93% (±1), relative to 47% (±1) for females mated to other males. Females more receptive to re-mating consistently produced fewer eggs. Females that produced many eggs had a lower receptivity, although in some cases there seemed to be an uncoupling of these 2 traits (Spearman rank correlation P-value = 0.001; Supplementary Figure S23).
Variation in Transcript Abundance After Mating is Correlated With Short-term Egg Production
Variation in post-mating transcript level changes significantly correlated with the number of eggs produced on day 1 after mating. No significant correlations were found with other phenotypes that we measured. Among the 77 genes that were differentially regulated depending on female × male genotype, levels of only one transcript, CG32277, correlated significantly with egg production. A more than 2-fold down-regulation of CG32277 in Beijing females mated to Beijing males was correlated with a higher day-one egg production (q = 0.03; Supplementary Figure S24). Virgin CPM values for CG32277 were similar across lines, but only Beijing females down-regulated CG32277 transcript levels after mating. CG32277 encodes a serine-type endopeptidase that is highly expressed in the spermathecae (Chintapalli et al. 2007). Among all 9484 genes in our dataset, the post-mating fold changes of 235 genes were significantly correlated with egg production on day 1 after mating (Supplementary File 3). Among the 235 genes, many have high expression in the ovary and known functions in oogenesis or ovulation, such as egg shell formation or octopamine signaling. In addition, the 235 genes included genes with GO terms related to metabolism, transcription and translation, cell division, nervous and muscle system processes, sensory perception and the immune response. Based on the phenotype data, females from the Beijing line produced the highest number of eggs on day 1, whereas females from the Netherlands line produced the lowest number of eggs. For the 235 genes, females from these 2 lines also differed most in their post-mating fold changes. For example, Beijing females underwent the strongest down-regulation of Tbh, a gene involved in ovulation (Monastirioti et al. 1996), while Netherlands females underwent the strongest up-regulation of Tbh. However, the differences in transcript-level fold changes between Beijing and Netherlands females were not significant.
Variation in Female Post-Mating Responses is not Correlated With Divergence Time Between the Female Genotype and the Genotype of her Mate
No clear correlation was found between the number or type of differentially regulated genes detected in our RNAseq data and whether the cross was intra- or inter-population. In intra-population crosses, an average of 2 differentially regulated genes were detected. In inter-population crosses, an average of 4.6 differentially regulated genes were found. However, the variance within each of these groups was large (Supplementary Table S3). Nevertheless, gene expression changes in Zimbabwe females never differed from the average. On the other hand, post-mating transcriptional changes in females from the Tasmania line were more prone to differ from the average due to female × male interactions (Supplementary Table S3). Likewise, no obvious correlations were found between variation in post-mating phenotypes and divergence time. Measures of fecundity were comparable between intra- and inter-population crosses, with an average of 32 (±7) eggs produced per day in an inter-population cross, and an average of 34 (±9) eggs produced per day in an intra-population cross (Figure 4B, Supplementary Figure S21). Females mated to males from their own population did not consistently demonstrate a higher refractoriness to re-mating (Figure 5B). We expected that hatchability would be higher in intra-population crosses, compared to inter-population crosses. However, this was not the case for the Beijing, Ithaca, and Tasmania lines (Figure 5A).
Discussion
In this study, we used natural variation in the Global Diversity Lines to assess female × male genotype interaction effects on egg production, hatchability, receptivity and female transcriptional responses to mating, in D. melanogaster. Significant female × male interactions were observed for all phenotypes measured here. Our RNAseq analysis identified molecules in females that might underlie female × male genotype-dependent variation affecting reproductive success.
Female × Male Genotypic Interactions Affect Post-Mating Responses in the Global Diversity Lines
Strong interactions between female and male genotype affect phenotypic post-mating responses in the Global Diversity Lines, consistent with previous observations in other lines (Chow et al. 2010; Giardina et al. 2011; Lüpold et al. 2013; Reinhart et al. 2015). The strongest effects in our study were observed for males from the Netherlands and Zimbabwe lines. These males were unable to induce long-term refractoriness to re-mating in all females. In addition, they failed to stimulate long-term egg production in multiple, but not all female backgrounds. Variation across the Global Diversity Lines in the male seminal fluid protein SP might underlie the observed phenotypic responses. SP is crucial for the initiation and maintenance of long-term post-mating responses (Chapman et al. 2003; Liu and Kubli 2003). Genetic variation might affect SP transfer, storage, or signaling (Cirera and Aguadé 1997; Chapman et al. 2003; Liu and Kubli 2003; Yapici et al. 2008; Chow et al. 2010; Smith et al. 2012). Refractoriness to re-mating was variable at 24h after the first mating. Shortly after mating, female receptivity is affected by seminal fluid proteins (Saudan et al. 2002; Chapman et al. 2003) and pheromones (Lebreton et al. 2014; Laturney and Billeter 2016). Because multiple factors contribute to short-term re-mating rate, stochastic variation in these factors could mask any female or male genotypic effects.
In addition to measuring post-mating phenotypes, we also measured post-mating transcript-level changes in the 24 mating combinations. Several studies have characterized post-mating changes in female transcript-abundance within one line (McGraw et al. 2004; Lawniczak and Begun 2004; Mack et al. 2006; Kapelnikov et al. 2008). We averaged post-mating gene expression changes across diverged lines, and found 272 differentially regulated genes. Sixty-one percent of these differentially expressed genes had previously been reported to respond to mating (Supplementary Files 1 and 2). These consistent gene expression changes might represent the essential transcriptional response to mating, instead of gene expression changes specific to one genetic background.
Female × male interactions at the transcript level would provide a mechanism that underlies the interactions that are observed at the phenotypic level. Unlike previous studies, we identified 77 genes whose change in post-mating transcript level deviated significantly from the average response to mating, depending on the combination of female and male genotype. McGraw et al. (2009) found negligible female × male interaction effects when using microarrays to measure post-mating transcriptional responses in whole females, 1–3 h after they mated with a male from their own strain or a male from a different inbred lab strain (Oregon R and Canton-S). Several explanations might account for the discrepancy between the prior and current study. First, shortly after mating, transcriptional changes may not occur because females are “poised” for reproduction, and males simply switch on proteins, RNAs, and molecules that are already present (McGraw et al. 2009). Given that our experiment identified female × male genotype effects at 5–6 h post-mating, interactions on the transcript level potentially occur after the 1–3 h window examined by McGraw et al. Additionally, stronger interactions might be induced by strains that are genetically more diverged (such as the Global Diversity Lines used here), compared to the 2 inbred lab strains used by McGraw et al. Antagonistic co-evolution of genes involved in these interactions could affect reproductive compatibility between diverged populations (e.g., Ting et al. 2001; Gavrilets 2014; Jennings et al. 2014; Sirot et al. 2015).
Transcripts That are Sensitive to Female × Male Genotypic Interactions Likely Underlie Variation in Phenotypic Post-Mating Responses, and Play a Role in Sexual Conflict
The 77 genes that are sensitive to female × male interactions likely point to mechanisms and biological processes that underlie variation in post-mating phenotypes, possibly through direct interaction with, or downstream responses to, male seminal fluid proteins, several of which have allelic variants known to cause alterations in phenotypic post-mating responses (Hughes 1997; Clark et al. 1999; Fiumera et al. 2005, 2006; Chow et al. 2010; Greenspan and Clark 2011; Lüpold et al. 2012; Zhang et al. 2013).
These 77 genes might act in a variety of tissues in order to mediate these responses. For example, variation in post-mating transcript levels of genes expressed in the spermathecae and their associated secretory cells could impact sperm storage (Lüpold et al. 2012, 2013) and maintenance (Schnakenberg et al. 2011; Sun and Spradling 2013). Differential regulation of genes expressed in the ovary could reflect a male’s capacity to induce egg production in a particular female background (Heifetz et al. 2001). Alterations in post-mating transcript levels of genes expressed in the digestive system potentially influence a female’s nutrient uptake and metabolism (Shingleton 2015), which might in turn influence female investment in egg production (Terashima et al. 2005). Differential expression of genes expressed in the head and/or genes that have sensory functions (vision and olfaction), could alter a female’s response to food or guide her to suitable oviposition sites (Matsuo et al. 2007; Harada et al. 2008; Gioti et al. 2012). Alternatively, differential expression of vision and olfaction-related genes could impact a female’s response to other females or males. Mating changes the abundance of both transcripts that encode odorant binding proteins, and odorant binding proteins themselves in females (McGraw et al. 2004; Findlay et al. 2008). Proper functioning of odorant binding proteins and odorant receptors is associated with female sensitivity to male pheromones and re-mating rate (Giardina et al. 2011; Lebreton et al. 2014). Snmp1, which is involved in the female response to the male pheromone 11-cis-vaccenyl acetate (Jin et al. 2008), was differentially regulated in our dataset depending on female and male genotype. This suggests that depending on female genotype, some male genotypes have stronger effects on female sensitivity to other males.
Female × male genotypic interactions also affected the expression of immune gene transcripts. Many studies report the up-regulation of immune gene expression after mating, in Drosophila (McGraw et al. 2004, 2008; Mack et al. 2006; Kapelnikov et al. 2008; Short and Lazzaro 2013), other insects (e.g., Baer et al. 2006; Shoemaker et al. 2006), and in vertebrates, including humans (Johansson et al. 2004; Robertson 2005; Richard et al. 2012; Schjenken and Robertson 2014). Our study is the first to observe that the intensity of this post-mating up-regulation of immune transcripts depends on an interaction between female and male genotype in D. melanogaster. Even though an immune response seems to be an inherent part of the post-mating response, whether it is adaptive in D. melanogaster remains speculative. The post-mating up-regulation of immune transcripts could prepare females to fight off sexually transmitted diseases. In this case, the response is beneficial for both sexes (Samakovlis et al. 1991; Lung et al. 2001; Lawniczak et al. 2007; Zhong et al. 2013). Alternatively, females might induce an up-regulation of AMPs after mating to compensate for the toxic effects of some seminal fluid proteins (Chapman et al. 1995; Wigby and Chapman 2005; Mueller et al. 2007; Innocenti and Morrow 2009; Morrow and Innocenti 2012). It is also possible that females employ the immune response to assess male quality or compatibility (Lawniczak et al. 2007).
The variation observed in the phenotypes and transcript levels described above could be the consequence of sexual conflict. In terms of the immune response, recently mated females have higher AMP mRNA levels, but they are less resistant to systemic bacterial infection than are virgin females, and this difference depends on the transfer of the ejaculate (Fedorka et al. 2007; Short et al. 2012). Genes that impact female olfactory behavior can alter female receptivity to future matings, while polyandry is thought to be beneficial for females and not males. Male influence on female egg production potentially causes sexual conflict as well. Males benefit if females produce many eggs shortly after mating, to ensure the female uses as much of the male’s sperm before mating with another male. On the other hand, females might suffer reduced lifetime reproductive output when investing many resources in egg production in a brief period of time (Sirot et al. 2015). We found that down-regulation of the spermathecal endopeptidase CG32277 in Beijing females correlated with a high day 1 egg production, a trait thought to benefit mainly males. Sexual conflict over CG32277 expression levels could have resulted in a transcriptional post-mating response that benefits males in the Beijing line. As CG32277 is a secreted peptidase, it has the opportunity to interact with male molecules transferred during mating, exposing CG32277 directly to pressures arising from sexual conflict. Similarly, Esp and CG8329 form potential targets of sexual conflict. Esp is a member of the “SP network,” a network of male and female proteins required to bind the male seminal fluid protein SP to sperm. This process is crucial to mediate long-term post-mating responses, including a long-term reduction in receptivity, in females (Findlay et al. 2014). Expression of CG8329 occurs both in the head (Chintapalli et al. 2007) and in spermathecae (Prokupek et al. 2009), and is regulated by the seminal proteins Acp62F and Acp29Ab at 1 to 3h after mating (McGraw et al. 2008).
Validation Using qRT-PCR
For 5 out of 6 genes, our RNAseq results were well validated using qRT-PCR. The exception was Obp49a, for which only one of 3 qRT-PCR replicates confirmed the findings from the RNAseq analysis. Although we did not dissect the causes for this, it is most likely due to unmeasured microenvironmental variation, such as might be caused by differences in the medium or microbial contamination.
Immune genes were up-regulated more than average in T × T and T × Z in the RNAseq dataset, but Tasmania virgins also had higher CPM values for a range of antimicrobial peptides (Supplementary Figure S17). This raised the concern that variation in immune gene transcripts post-mating was caused by infection rather than mating. Validation using a separate QRT-PCR assay was necessary to ensure that the observed results in T × T and T × Z were reproducible, and were not due to the presence of pathogens in the Tasmania stocks used for RNAseq sample collection. The up-regulation of Def in T × T was validated using qRT-PCR, and mRNA levels for antimicrobial peptides were not found to be higher in Tasmania virgins relative to Beijing and Ithaca virgins in the qRT-PCR assays. This suggests that the observed up-regulation of Def is in fact due to genotypic interactions.
Female × Male Genotypic Interactions are More Prevalent Than Male- or Female-Genotype Dependent Effects
No significant differences were found in the transcriptional response to mating depending on male genotype alone. A probable reason is that the role of the male is thought to be limited to triggering the post-mating response. Once the switch from “unmated” to “mated” has been made in the female (and this switch occurs before 5–6 h after mating), robust female responses take over (Heifetz and Wolfner 2004; Heifetz et al. 2014; Heifetz and Wolfner 2004; Mattei et al. 2015; Carmel et al. 2016). Additionally, male-only genotype effects might be rare, as the effect of variable seminal fluid protein composition across diverged males would also depend on female genotype-specific sensitivity to this variation in protein composition.
Genes differently regulated depending on only the female genotype (and averaged across male genotypes) were only found in females from the Tasmania line. These differentially regulated genes encoded among others proteases and digestive system-specific proteins, suggesting that post-mating metabolism differs by female genotype. Additionally, the differential regulation of 2 genes encoding proteins involved in the perception of taste, suggests that female genotypes vary in post-mating gustatory processes. Mating affects these processes, presumably to enhance nutrient intake and reproductive output (Walker et al. 2015). In general, females from the diverged, geographically isolated populations used here did not show drastic differences in their response to mating. Mating causes systemic changes that affect complex, polygenic traits (McGraw et al. 2004; Carvalho et al. 2006; Cognigni et al. 2011; Apger-McGlaughon and Wolfner 2013; Short and Lazzaro 2013; Reiff et al. 2015). Females from different populations might differ slightly in the timing of regulation of the genes involved, or might favor one gene over the other if there is redundancy. Overall, transcriptional changes caused by mating appear to be robust with respect to female genotype.
Variation in Female Post-Mating Responses is not Correlated With Divergence Time Between Female and Male Genotypes of the Global Diversity Lines
We expected to see evidence of co-evolution in gene expression patterns, where inter-population crosses, or crosses between more diverged lines would lead to stronger effects on post-mating phenotypes. For example, in ants, queens mated to an allopatric male had higher levels of immune response mRNAs, but this did not happen if the queen had mated with a sympatric male (Schrempf et al. 2015). Among the 5 lines used in this study, the lines from Ithaca and Tasmania diverged more recently than the lines from Beijing and the Netherlands (Grenier et al. 2015). Drosophila melanogaster originated in Africa (Lachaise et al. 1988), but the line from Zimbabwe used here was a recent migrant (Grenier et al. 2015). In our study, we did not see correlations between the strength of post-mating responses and divergence time. We observed, however, that females from distinct lines differed in their sensitivity to female × male genotypic interactions. This suggests that at the transcriptome level, females from isolated populations differ in how sensitive they are to male input. In addition, the timing of female × male genotype-dependent transcriptional responses might vary across females from isolated populations. Finally, we detected low intra-population hatchability in the Beijing, Tasmania and Ithaca lines, likely due to some degree of inbreeding depression. Kao et al. (2015) have suggested that low hatchability due to hybrid incompatibility could contribute to incipient speciation between D. melanogaster lines, but we saw no evidence for hybrid incompatibility among the lines we tested.
Conclusions
Our results demonstrate the widespread effects of female × male genotypic interactions in processes related to reproduction, going from post-mating transcriptional responses to physiological and behavioral changes. Genes affected by female × male genotypic interactions included ones involved in the immune response, ones that might impact egg laying, and ones that are likely involved in post-mating behavioral changes. Future work is needed to uncover the precise roles these genes play in the female post-mating response, to determine to what extent these genes are indicators of female × male compatibility on a molecular level, and to determine if they are affected by (antagonistic) sexual selection.
Supplementary Material
Supplementary data are available at Journal of Heredity online.
Funding
This work was supported in part by the National Institutes of Health (R01–HD059060 to A.G.C. and M.F.W.). For part of this work, S.Y.N.D. was supported by a fellowship from the Belgian American Educational Foundation and C.Y.C. was supported by a traineeship from the National Institutes of Health (training grant T32-HD052471).
Data Availability
The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE104706, http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE104706.
Supplementary Material
Acknowledgments
We thank E. J. Cosgrove, L. M. Johnson (CSCU), J. Grenier, J. Pleiss, Y. Ahmed-Braimah, Y. Hafezi, and E. Davenport for helpful discussions and advice on statistical analyses, A. Manfredo for the RNA-seq library preparation, Z. Waseem for help with fecundity and hatchability assays, and S. Wigby and anonymous reviewers for the helpful comments and suggestions.
References
- Anders S, Pyl PT, Huber W. 2015. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 31:166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrés JA, Arnqvist G. 2001. Genetic divergence of the seminal signal-receptor system in houseflies: the footprints of sexually antagonistic coevolution?Proc Biol Sci. 268:399–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apger-McGlaughon J, Wolfner MF. 2013. Post-mating change in excretion by mated Drosophila melanogaster females is a long-term response that depends on sex peptide and sperm. J Insect Physiol. 59:1024–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attrill H, Falls K, Goodman JL, Millburn GH, Antonazzo G, Rey AJ, Marygold SJ; FlyBase Consortium 2016. FlyBase: establishing a Gene Group resource for Drosophila melanogaster. Nucleic Acids Res. 44:D786–D792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avila FW, Sirot LK, LaFlamme BA, Rubinstein CD, Wolfner MF. 2011. Insect seminal fluid proteins: identification and function. Annu Rev Entomol. 56:21–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer B, Armitage SA, Boomsma JJ. 2006. Sperm storage induces an immunity cost in ants. Nature. 441:872–875. [DOI] [PubMed] [Google Scholar]
- Benjamini Y.and Hochberg Y. 1995. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society series B. 57:289–300. [Google Scholar]
- Birkhead T.R., Pizzari T. 2002. Post-copulatory sexual selection. Nat. rev. Genet . 3:262–273. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bono JM, Matzkin LM, Kelleher ES, Markow TA. 2011. Postmating transcriptional changes in reproductive tracts of con- and heterospecifically mated Drosophila mojavensis females. Proc Natl Acad Sci U S A. 108:7878–7883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carhan A, Tang K, Shirras CA, Shirras AD, Isaac RE. 2011. Loss of Angiotensin-converting enzyme-related (ACER) peptidase disrupts night-time sleep in adult Drosophila melanogaster. J Exp Biol. 214:680–686. [DOI] [PubMed] [Google Scholar]
- Carmel I, Tram U, Heifetz Y. 2016. Mating induces developmental changes in the insect female reproductive tract. Curr Opin Insect Sci. 13:106–113. [DOI] [PubMed] [Google Scholar]
- Carvalho GB, Kapahi P, Anderson DJ, Benzer S. 2006. Allocrine modulation of feeding behavior by the sex peptide of Drosophila. Curr Biol. 16:692–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman T, Bangham J, Vinti G, Seifried B, Lung O, Wolfner MF, Smith HK, Partridge L. 2003. The sex peptide of Drosophila melanogaster: female post-mating responses analyzed by using RNA interference. Proc Natl Acad Sci U S A. 100:9923–9928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman T, Liddle LF, Kalb JM, Wolfner MF, Partridge L. 1995. Cost of mating in Drosophila melanogaster females is mediated by male accessory gland products. Nature. 373:241–244. [DOI] [PubMed] [Google Scholar]
- Chintapalli VR, Wang J, Dow JA. 2007. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet. 39:715–720. [DOI] [PubMed] [Google Scholar]
- Chow CY, Wolfner MF, Clark AG. 2010. The genetic basis for male x female interactions underlying variation in reproductive phenotypes of Drosophila. Genetics. 186:1355–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow CY, Wolfner MF, Clark AG. 2013. Large neurological component to genetic differences underlying biased sperm use in Drosophila. Genetics. 193:177–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirera S, Aguadé M. 1997. Evolutionary history of the sex-peptide (Acp70A) gene region in Drosophila melanogaster. Genetics. 147:189–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AG, Aguadé M, Prout T, Harshman LG, Langley CH. 1995. Variation in sperm displacement and its association with accessory gland protein loci in Drosophila melanogaster. Genetics. 139:189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AG, Begun DJ. 1998. Female genotypes affect sperm displacement in Drosophila. Genetics. 149:1487–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AG, Begun DJ, Prout T. 1999. Female x male interactions in Drosophila sperm competition. Science. 283:217–220. [DOI] [PubMed] [Google Scholar]
- Clark AG, Dermitzakis ET, Civetta A. 2000. Nontransitivity of sperm precedence in Drosophila. Evolution. 54:1030–1035. [DOI] [PubMed] [Google Scholar]
- Cognigni P, Bailey AP, Miguel-Aliaga I. 2011. Enteric neurons and systemic signals couple nutritional and reproductive status with intestinal homeostasis. Cell Metab. 13:92–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton JE, Kacheria TS, Knott SR, Lebo MS, Nishitani A, Sanders LE, Stirling EJ, Winbush A, Arbeitman MN. 2010. Dynamic, mating-induced gene expression changes in female head and brain tissues of Drosophila melanogaster. BMC Genomics. 11:541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domanitskaya EV, Liu H, Chen S, Kubli E. 2007. The hydroxyproline motif of male sex peptide elicits the innate immune response in Drosophila females. FEBS J. 274:5659–5668. [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30:207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorka KM, Linder JE, Winterhalter W, Promislow D. 2007. Post-mating disparity between potential and realized immune response in Drosophila melanogaster. Proc Biol Sci. 274:1211–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findlay GD, Sitnik JL, Wang W, Aquadro CF, Clark NL, Wolfner MF. 2014. Evolutionary rate covariation identifies new members of a protein network required for Drosophila melanogaster female post-mating responses. PLoS Genet. 10:e1004108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findlay GD, Yi X, Maccoss MJ, Swanson WJ. 2008. Proteomics reveals novel Drosophila seminal fluid proteins transferred at mating. PLoS Biol. 6:e178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiumera AC, Dumont BL, Clark AG. 2005. Sperm competitive ability in Drosophila melanogaster associated with variation in male reproductive proteins. Genetics. 169:243–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiumera AC, Dumont BL, Clark AG. 2006. Natural variation in male-induced ‘cost-of-mating’ and allele-specific association with male reproductive genes in Drosophila melanogaster. Philos Trans R Soc Lond B Biol Sci. 361:355–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrilets S. 2014. Is sexual conflict an “engine of speciation”?Cold Spring Harb Perspect Biol. 6:a017723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardina TJ, Beavis A, Clark AG, Fiumera AC. 2011. Female influence on pre- and post-copulatory sexual selection and its genetic basis in Drosophila melanogaster. Mol Ecol. 20:4098–4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson G. 2008. The environmental contribution to gene expression profiles. Nat Rev Genet. 9:575–581. [DOI] [PubMed] [Google Scholar]
- Gioti A, Wigby S, Wertheim B, Schuster E, Martinez P, Pennington CJ, Partridge L, Chapman T. 2012. Sex peptide of Drosophila melanogaster males is a global regulator of reproductive processes in females. Proc Biol Sci. 279:4423–4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenspan L, Clark AG. 2011. Associations between variation in X chromosome male reproductive genes and sperm competitive ability in Drosophila melanogaster. Int J Evol Biol. 2011:214280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenier JK, Arguello JR, Moreira MC, Gottipati S, Mohammed J, Hackett SR, Boughton R, Greenberg AJ, Clark AG. 2015. Global diversity lines - a five-continent reference panel of sequenced Drosophila melanogaster strains. G3 (Bethesda). 5:593–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada E, Haba D, Aigaki T, Matsuo T. 2008. Behavioral analyses of mutants for two odorant-binding protein genes, Obp57d and Obp57e, in Drosophila melanogaster. Genes Genet Syst. 83:257–264. [DOI] [PubMed] [Google Scholar]
- Heifetz Y, Lindner M, Garini Y, Wolfner MF. 2014. Mating regulates neuromodulator ensembles at nerve termini innervating the Drosophila reproductive tract. Curr Biol. 24:731–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heifetz Y, Tram U, Wolfner MF. 2001. Male contributions to egg production: the role of accessory gland products and sperm in Drosophila melanogaster. Proc Biol Sci. 268:175–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heifetz Y, Wolfner MF. 2004. Mating, seminal fluid components, and sperm cause changes in vesicle release in the Drosophila female reproductive tract. Proc Natl Acad Sci U S A. 101:6261–6266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann AA, Hercus M, Dagher H. 1998. Population dynamics of the Wolbachia infection causing cytoplasmic incompatibility in Drosophila melanogaster. Genetics. 148:221–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollis B, Houle D, Kawecki TJ. 2016. Evolution of reduced post-copulatory molecular interactions in Drosophila populations lacking sperm competition. J Evol Biol. 29:77–85. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. 2008. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 4:44–57. [DOI] [PubMed] [Google Scholar]
- Huang DAW, Sherman BT, Lempicki RA. 2009. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes KA. 1997. Quantitative genetics of sperm precedence in Drosophila melanogaster. Genetics. 145:139–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innocenti P, Morrow EH. 2009. Immunogenic males: a genome-wide analysis of reproduction and the cost of mating in Drosophila melanogaster females. J Evol Biol. 22:964–973. [DOI] [PubMed] [Google Scholar]
- Jennings JH, Snook RR, Hoikkala A. 2014. Reproductive isolation among allopatric Drosophila montana populations. Evolution. 68:3095–3108. [DOI] [PubMed] [Google Scholar]
- Jin X, Ha TS, Smith DP. 2008. SNMP is a signaling component required for pheromone sensitivity in Drosophila. Proc Natl Acad Sci U S A. 105:10996–11001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M, Bromfield JJ, Jasper MJ, Robertson SA. 2004. Semen activates the female immune response during early pregnancy in mice. Immunology. 112:290–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao JY, Lymer S, Hwang SH, Sung A, Nuzhdin SV. 2015. Postmating reproductive barriers contribute to the incipient sexual isolation of the United States and Caribbean Drosophila melanogaster. Ecol Evol. 5:3171–3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapelnikov A, Rivlin PK, Hoy RR, Heifetz Y. 2008. Tissue remodeling: a mating-induced differentiation program for the Drosophila oviduct. BMC Dev Biol. 8:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick M, Ravigné V. 2002. Speciation by natural and sexual selection: models and experiments. Am Nat. 159(Suppl 3):S22–35. [DOI] [PubMed] [Google Scholar]
- Lachaise D., Cariou M.-L., David J.R., Lemeunier F., Tsacas L., and Ashburner M. 1988. Historical Biogeography of the Drosophila melanogaster Species Subgroup. In Evolutionary Biology, M.K. Hecht, B. Wallace, and G.T. Prance, eds. (Springer US), pp. 159–225. [Google Scholar]
- Laturney M, Billeter JC. 2016. Drosophila melanogaster females restore their attractiveness after mating by removing male anti-aphrodisiac pheromones. Nat Commun. 7:12322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawniczak MK, Barnes AI, Linklater JR, Boone JM, Wigby S, Chapman T. 2007. Mating and immunity in invertebrates. Trends Ecol Evol. 22:48–55. [DOI] [PubMed] [Google Scholar]
- Lawniczak MK, Begun DJ. 2004. A genome-wide analysis of courting and mating responses in Drosophila melanogaster females. Genome. 47:900–910. [DOI] [PubMed] [Google Scholar]
- Lawniczak MK, Begun DJ. 2005. A QTL analysis of female variation contributing to refractoriness and sperm competition in Drosophila melanogaster. Genet Res. 86:107–114. [DOI] [PubMed] [Google Scholar]
- Lebreton S, Grabe V, Omondi AB, Ignell R, Becher PG, Hansson BS, Sachse S, Witzgall P. 2014. Love makes smell blind: mating suppresses pheromone attraction in Drosophila females via Or65a olfactory neurons. Sci Rep. 4:7119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenth R.V. 2016. Least Square Means: The R package lsmeans. Journal of Statistical Software. 69” No page numbers are available for this reference, but this is the url: https://www.jstatsoft.org/article/view/v069i01 [Google Scholar]
- Liu H, Kubli E. 2003. Sex-peptide is the molecular basis of the sperm effect in Drosophila melanogaster. Proc Natl Acad Sci U S A. 100:9929–9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. [DOI] [PubMed] [Google Scholar]
- Lung O, Kuo L, Wolfner MF. 2001. Drosophila males transfer antibacterial proteins from their accessory gland and ejaculatory duct to their mates. J Insect Physiol. 47:617–622. [DOI] [PubMed] [Google Scholar]
- Lüpold S, Manier MK, Berben KS, Smith KJ, Daley BD, Buckley SH, Belote JM, Pitnick S. 2012. How multivariate ejaculate traits determine competitive fertilization success in Drosophila melanogaster. Curr Biol. 22:1667–1672. [DOI] [PubMed] [Google Scholar]
- Lüpold S, Pitnick S, Berben KS, Blengini CS, Belote JM, Manier MK. 2013. Female mediation of competitive fertilization success in Drosophila melanogaster. Proc Natl Acad Sci U S A. 110:10693–10698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack PD, Kapelnikov A, Heifetz Y, Bender M. 2006. Mating-responsive genes in reproductive tissues of female Drosophila melanogaster. Proc Natl Acad Sci U S A. 103:10358–10363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magezi DA. 2015. Linear mixed-effects models for within-participant psychology experiments: an introductory tutorial and free, graphical user interface (LMMgui). Front Psychol. 6:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo T, Sugaya S, Yasukawa J, Aigaki T, Fuyama Y. 2007. Odorant-binding proteins OBP57d and OBP57e affect taste perception and host-plant preference in Drosophila sechellia. PLoS Biol. 5:e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattei AL, Riccio ML, Avila FW, Wolfner MF. 2015. Integrated 3D view of postmating responses by the Drosophila melanogaster female reproductive tract, obtained by micro-computed tomography scanning. Proc Natl Acad Sci U S A. 112:8475–8480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy DJ, Chen Y, Smyth GK. 2012. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 40:4288–4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGraw LA, Clark AG, Wolfner MF. 2008. Post-mating gene expression profiles of female Drosophila melanogaster in response to time and to four male accessory gland proteins. Genetics. 179:1395–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGraw LA, Gibson G, Clark AG, Wolfner MF. 2009. Strain-dependent differences in several reproductive traits are not accompanied by early postmating transcriptome changes in female Drosophila melanogaster. Genetics. 181:1273–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGraw LA, Gibson G, Clark AG, Wolfner MF. 2004. Genes regulated by mating, sperm, or seminal proteins in mated female Drosophila melanogaster. Curr Biol. 14:1509–1514. [DOI] [PubMed] [Google Scholar]
- Monastirioti M, Linn CE Jr, White K. 1996. Characterization of Drosophila tyramine beta-hydroxylase gene and isolation of mutant flies lacking octopamine. J Neurosci. 16:3900–3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow EH, Innocenti P. 2012. Female postmating immune responses, immune system evolution and immunogenic males. Biol Rev Camb Philos Soc. 87:631–638. [DOI] [PubMed] [Google Scholar]
- Mueller JL, Page JL, Wolfner MF. 2007. An ectopic expression screen reveals the protective and toxic effects of Drosophila seminal fluid proteins. Genetics. 175:777–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson T, Fricke C, Arnqvist G. 2003. The effects of male and female genotype on variance in male fertilization success in the red flour beetle (Tribolium castaneum). Behav Ecol Sociobiol. 53:227–233. [Google Scholar]
- Panhuis TM, Butlin R, Zuk M, Tregenza T. 2001. Sexual selection and speciation. Trends Ecol Evol. 16:364–371. [DOI] [PubMed] [Google Scholar]
- Panhuis TM, Swanson WJ. 2006. Molecular evolution and population genetic analysis of candidate female reproductive genes in Drosophila. Genetics. 173:2039–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phadnis N, Orr HA. 2009. A single gene causes both male sterility and segregation distortion in Drosophila hybrids. Science. 323:376–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poinsot D, Bourtzis K, Markakis G, Savakis C, Merçot H. 1998. Wolbachia transfer from Drosophila melanogaster into D. simulans: host effect and cytoplasmic incompatibility relationships. Genetics. 150:227–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponton F, Chapuis MP, Pernice M, Sword GA, Simpson SJ. 2011. Evaluation of potential reference genes for reverse transcription-qPCR studies of physiological responses in Drosophila melanogaster. J Insect Physiol. 57:840–850. [DOI] [PubMed] [Google Scholar]
- Prokupek AM, Kachman SD, Ladunga I, Harshman LG. 2009. Transcriptional profiling of the sperm storage organs of Drosophila melanogaster. Insect Mol Biol. 18:465–475. [DOI] [PubMed] [Google Scholar]
- Prout T, Clark AG. 1996. Polymorphism in genes that influence sperm displacement. Genetics. 144:401–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiff T, Jacobson J, Cognigni P, Antonello Z, Ballesta E, Tan KJ, Yew JY, Dominguez M, Miguel-Aliaga I. 2015. Endocrine remodelling of the adult intestine sustains reproduction in Drosophila. Elife. 4:e06930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart M, Carney T, Clark AG, Fiumera AC. 2015. Characterizing male-female interactions using natural genetic variation in Drosophila melanogaster. J Hered. 106:67–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard M, Massot M, Clobert J, Meylan S. 2012. Litter quality and inflammatory response are dependent on mating strategy in a reptile. Oecologia. 170:39–46. [DOI] [PubMed] [Google Scholar]
- Rio DC, Ares M Jr, Hannon GJ, Nilsen TW. 2010. Purification of RNA using TRIzol (TRI reagent). Cold Spring Harb Protoc. 2010:pdb.prot5439. [DOI] [PubMed] [Google Scholar]
- Ritchie MG. 2007. Sexual selection and speciation. Annu Rev Ecol Evol Syst. 38, 79–102. [Google Scholar]
- Robertson SA. 2005. Seminal plasma and male factor signalling in the female reproductive tract. Cell Tissue Res. 322:43–52. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samakovlis C, Kylsten P, Kimbrell DA, Engström A, Hultmark D. 1991. The andropin gene and its product, a male-specific antibacterial peptide in Drosophila melanogaster. EMBO J. 10:163–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satyaki PR, Cuykendall TN, Wei KH, Brideau NJ, Kwak H, Aruna S, Ferree PM, Ji S, Barbash DA. 2014. The Hmr and Lhr hybrid incompatibility genes suppress a broad range of heterochromatic repeats. PLoS Genet. 10:e1004240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saudan P, Hauck K, Soller M, Choffat Y, Ottiger M, Spörri M, Ding Z, Hess D, Gehrig PM, Klauser S et al. . 2002. Ductus ejaculatorius peptide 99B (DUP99B), a novel Drosophila melanogaster sex-peptide pheromone. Eur J Biochem. 269:989–997. [DOI] [PubMed] [Google Scholar]
- Schjenken JE, Robertson SA. 2014. Seminal fluid and immune adaptation for pregnancy--comparative biology in mammalian species. Reprod. Domest Anim Zuchthyg. 49(Suppl 3):27–36. [DOI] [PubMed] [Google Scholar]
- Schnakenberg SL, Matias WR, Siegal ML. 2011. Sperm-storage defects and live birth in Drosophila females lacking spermathecal secretory cells. PLoS Biol. 9:e1001192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrempf A, von Wyschetzki K, Klein A, Schrader L, Oettler J, Heinze J. 2015. Mating with an allopatric male triggers immune response and decreases longevity of ant queens. Mol Ecol. 24:3618–3627. [DOI] [PubMed] [Google Scholar]
- Shingleton AW. 2015. Physiology: female flies have the guts for reproduction. Curr Biol. 25:R716–R718. [DOI] [PubMed] [Google Scholar]
- Shoemaker KL, Parsons NM, Adamo SA. 2006. Mating enhances parasite resistance in the cricket Gryllus texensis. Anim Behav. 71, 371–380. [Google Scholar]
- Short SM, Lazzaro BP. 2013. Reproductive status alters transcriptomic response to infection in female Drosophila melanogaster. G3 (Bethesda). 3:827–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short SM, Wolfner MF, Lazzaro BP. 2012. Female Drosophila melanogaster suffer reduced defense against infection due to seminal fluid components. J Insect Physiol. 58:1192–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirot LK, Wong A, Chapman T, Wolfner MF. 2015. Sexual conflict and seminal fluid proteins: a dynamic landscape of sexual interactions. Cold Spring Harb Perspect Biol. 7:a017533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DT, Sirot LK, Wolfner MF, Hosken DJ, Wedell N. 2012. The consequences of genetic variation in sex peptide expression levels for egg laying and retention in females. Heredity (Edinb). 109:222–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Spradling AC. 2013. Ovulation in Drosophila is controlled by secretory cells of the female reproductive tract. Elife. 2:e00415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson WJ, Clark AG, Waldrip-Dail HM, Wolfner MF, Aquadro CF. 2001. Evolutionary EST analysis identifies rapidly evolving male reproductive proteins in Drosophila. Proc Natl Acad Sci U S A. 98:7375–7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson WJ, Vacquier VD. 2002. The rapid evolution of reproductive proteins. Nat Rev Genet. 3:137–144. [DOI] [PubMed] [Google Scholar]
- Swanson WJ, Wong A, Wolfner MF, Aquadro CF. 2004. Evolutionary expressed sequence tag analysis of Drosophila female reproductive tracts identifies genes subjected to positive selection. Genetics. 168:1457–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S, Presgraves DC. 2015. Lineage-Specific Evolution of the Complex Nup160 Hybrid Incompatibility Between Drosophila melanogaster and Its Sister Species. Genetics. 200:1245–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terashima J, Takaki K, Sakurai S, Bownes M. 2005. Nutritional status affects 20-hydroxyecdysone concentration and progression of oogenesis in Drosophila melanogaster. J Endocrinol. 187:69–79. [DOI] [PubMed] [Google Scholar]
- Theopold U, Rissler M, Fabbri M, Schmidt O, Natori S. 1999. Insect glycobiology: a lectin multigene family in Drosophila melanogaster. Biochem Biophys Res Commun. 261:923–927. [DOI] [PubMed] [Google Scholar]
- Ting CT, Takahashi A, Wu CI. 2001. Incipient speciation by sexual isolation in Drosophila: concurrent evolution at multiple loci. Proc Natl Acad Sci U S A. 98:6709–6713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 7:562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker SJ, Corrales-Carvajal VM, Ribeiro C. 2015. Postmating circuitry modulates salt taste processing to increase reproductive output in Drosophila. Curr Biol CB. 25:2621–2630. [DOI] [PubMed] [Google Scholar]
- Wigby S, Chapman T. 2005. Sex peptide causes mating costs in female Drosophila melanogaster. Curr Biol. 15:316–321. [DOI] [PubMed] [Google Scholar]
- Yapici N, Kim YJ, Ribeiro C, Dickson BJ. 2008. A receptor that mediates the post-mating switch in Drosophila reproductive behaviour. Nature. 451:33–37. [DOI] [PubMed] [Google Scholar]
- Zhang R, Clark AG, Fiumera AC. 2013. Natural genetic variation in male reproductive genes contributes to nontransitivity of sperm competitive ability in Drosophila melanogaster. Mol Ecol. 22:1400–1415. [DOI] [PubMed] [Google Scholar]
- Zhong W, McClure CD, Evans CR, Mlynski DT, Immonen E, Ritchie MG, Priest NK. 2013. Immune anticipation of mating in Drosophila: Turandot M promotes immunity against sexually transmitted fungal infections. Proc Biol Sci. 280:20132018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S, Mackay T, Anholt RR. 2014. Transcriptional and epigenetic responses to mating and aging in Drosophila melanogaster. BMC Genomics. 15:927. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE104706, http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE104706.