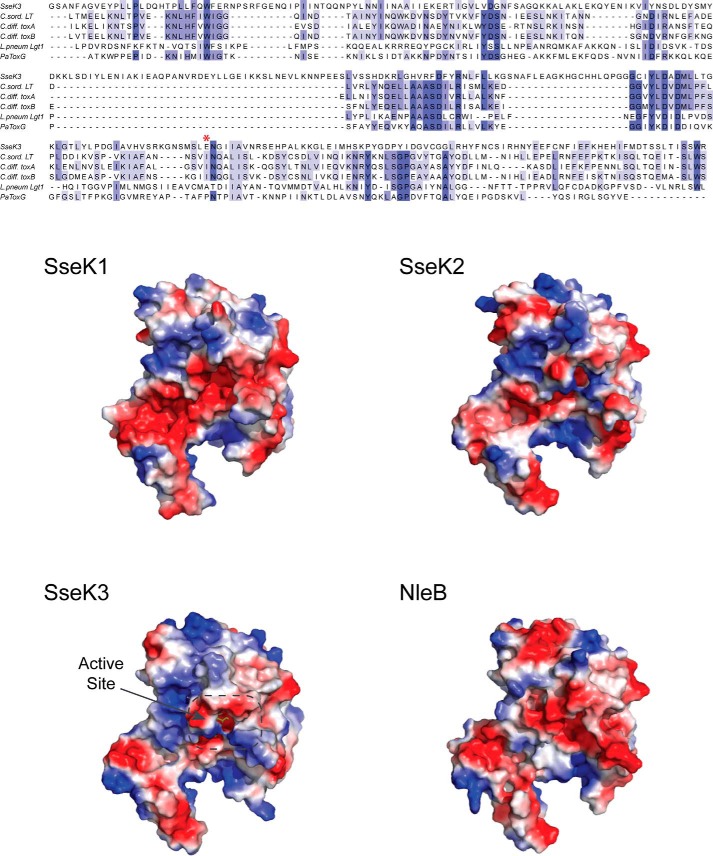
Figure 6.
Glycosyltransferase structural alignment and SseK1, SseK2, and NleB1 homology structural models. Top, structural alignment of SseK3 with C. sordelli LT (PDB code 2VKD, 19% sequence identity), C. difficile toxin A (3SRZ, 17%), toxin B (2BVL, 15%), L. pneumophila Lgt1 (3JSZ, 11%), and P. asymbiotica toxin (4MIX, 19%). The catalytically important Glu-258 residue is marked with an asterisk in the alignment. Bottom, solvent-accessible surface representation colored according to the electrostatic potential (blue, positive; red, negative) of the structure of SseK3(14–335) bound to its UDP-GlcNAc ligand and homology structural models of SseK1 (55% sequence identity with SseK3), Ssek2 (72%), and NleB1 (53%). Ligand bound in the active site is highlighted.