Figure 2.
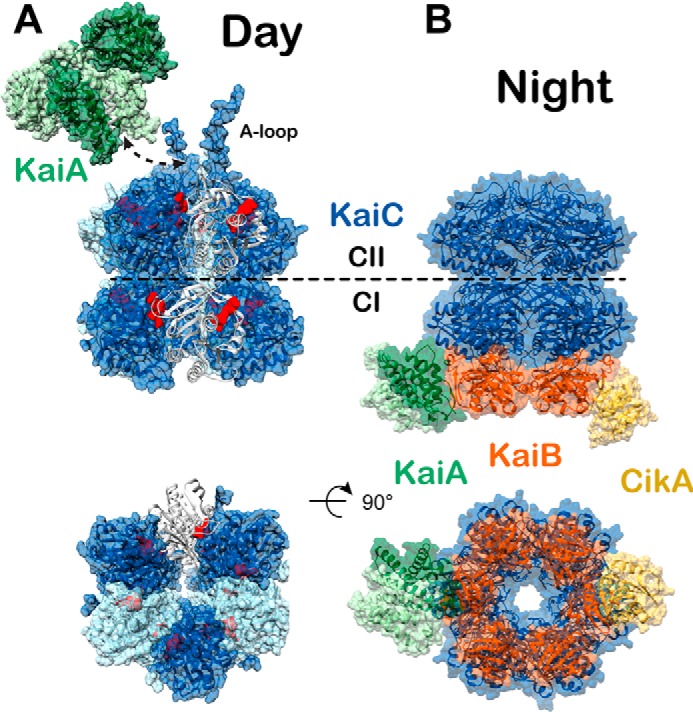
Day and night states of the cyanobacterial clock. High-resolution models of the day and nighttime states of the core oscillator are compared. Coloring scheme is the same as in Fig. 1. A, day complex. The CI and CII hexamers form stacked doughnuts (alternating subunits shown in light and dark blue for contrast, PDB code 3K0C). One subunit is shown in cartoon mode to highlight the nucleotide-binding sites (red) at the subunit interfaces. Phosphorylation of KaiC residues Thr-426 and Ser-432 takes place near the CII nucleotide-binding sites. The KaiA dimer (light and dark green for contrast) binds to C-terminal A-loops of KaiC (white, shown in complex with KaiA PDB code 5C5E). The dashed arrow represents the point of connection between the KaiA–CII loop structure and the CII A-loop extensions shown on the right. B, night complex. The intermediate-resolution cryo-EM model (PDB code 5N8Y) is combined with higher resolution models from studies on independent subcomplexes. The S431E phosphomimetic of the pS/T state of KaiC binds six molecules of KaiB (PDB code 5JWQ). This results in the recruitment and sequestration of KaiA near the KaiB–CI interface (PDB code 5JWR). Output signaling occurs through interactions between KaiB and the C-terminal PsR domain of CikA (PDB code 5JYV), as well as through interactions between KaiC and SasA at dusk, although no high-resolution structure exists for the latter.
