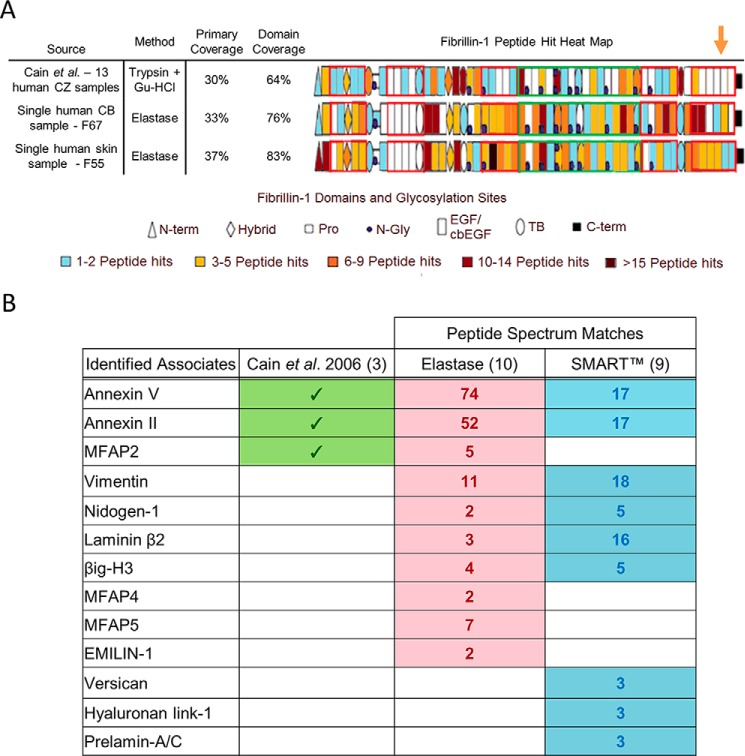
Figure 1.
Elastase and SMARTTM methods led to the improved detection of fibrillin-1 and improved identification of microfibril-associated proteins compared with previous published efforts. The ability of the elastase method to produce peptides of fibrillin-1 from a single human CB sample (female age 67; F67) and a single human skin sample (F55) is compared with the efforts of Cain et al. (23) (A). As performed by Cain et al. (23), PSMs (Peptide Prophet FDR ≤ 5%) were counted for each respective fibrillin-1 domain and heat-mapped. Our method led to a greater primary coverage and domain coverage from a single sample run than Cain et al. (23), whose coverages were achieved from 13 separate sample runs. Peptides from the C-terminal region of fibrillin-1 were also successfully detected (orange arrow), which Cain et al. (23) failed to identify. The known fibrillin microfibril-interacting proteins identified by Cain et al. (23) (green) are compared with those identified by elastase and SMARTTM methods (Protein Prophet FDR ≤ 0.1%, Peptide Prophet FDR ≤ 5%) (B). The elastase method (red) and SMARTTM method (blue) were both performed on the same human CB microfibril extract (F73). The elastase method appears to enhance the detection of microfibril-associated proteins thought to be tightly bonded to the structure (i.e. the MFAP family), whereas the SMARTTM method appears to enhance the detection of weakly interacting proteins (i.e. versican and hyaluronan proteins). Collectively, these methods led to an enhanced detection of known microfibril-associated proteins compared with Cain et al. (23).