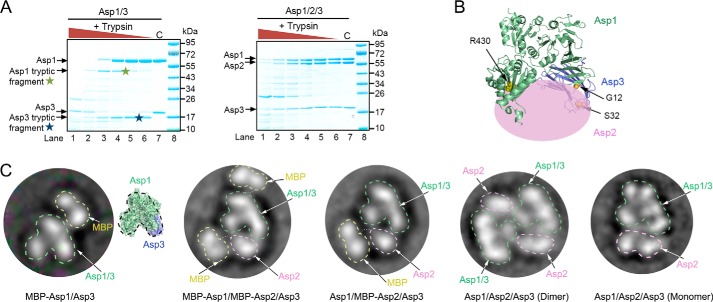
Figure 5.
Domain organization of the Asp1/2/3 complex. A, purified Asp1/3 and Asp1/2/3 complexes were incubated with different concentrations of trypsin. The samples were analyzed by SDS-PAGE and staining with Coomassie Blue. Tryptic fragments seen only with Asp1/3 are indicated by stars. C, control without trypsin. B, approximate position of Asp2 in the Asp1/2/3 complex. The crystal structure of the Asp1/3 complex is shown as a ribbon diagram and Asp2 as a pink ellipse. Trypsin cleavage at residues Arg-430 and Arg-23 is prevented by Asp2. R430 is indicated as yellow balls. Arg-23 is located in a flexible region that is invisible in the crystal structure, so residues G12 and S32 in the flanking segments are shown (in orange). C, left three panels show the domain organization of Asp complexes that contain fusions of MBP to Asp1 and/or Asp2. Shown are 2D averages of particles analyzed by negative-stain electron microscopy. The MBP tag prevents dimerization of the Asp1/2/3 complex. The characteristic two-globule shape of MBP and the domain structure of Asp1/3, as deduced from a comparison with the crystal structure, were used to identify the proteins. The right two panels show the domain organization of dimeric and monomeric untagged Asp1/2/3 complex.