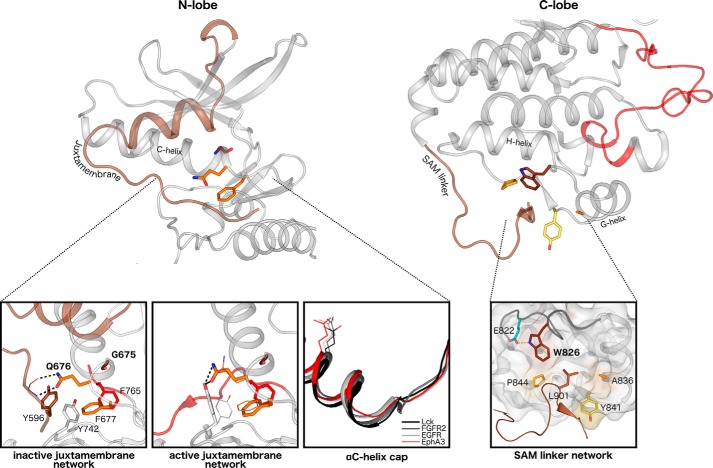
Figure 3.
Structural interactions in the juxtamembrane network and SAM domain linker network. Left, key interactions among the juxtamembrane, GQF motif, and kinase domain residues are highlighted in the autoinhibited (Protein Data Bank code 2QO2) and active (Protein Data Bank code 2QOC) structures of EphA3. The juxtamembrane tyrosine is shown in brown, the tethering GQF motif is in orange, and the DFG phenylalanine is in red. Different side chain occupancies of residues observed in the active structure are shown as lines. An alignment of αC helices of various tyrosine kinases reveals the earlier helix capping observed in Eph structures. The conserved ATP-binding αC helix glutamate is shown as a reference for the structural alignment. Right, interactions between the SAM domain linker and the C-lobe of the kinase domain are highlighted in an active structure of EphA3 (Protein Data Bank code 2QOC). The hydrophobic pocket in the C-lobe is depicted in surface representation, and the surface of Eph-specific residues that make up the pocket is colored. The αF-αG loop backbone is highlighted in dark gray.