Figure 6.
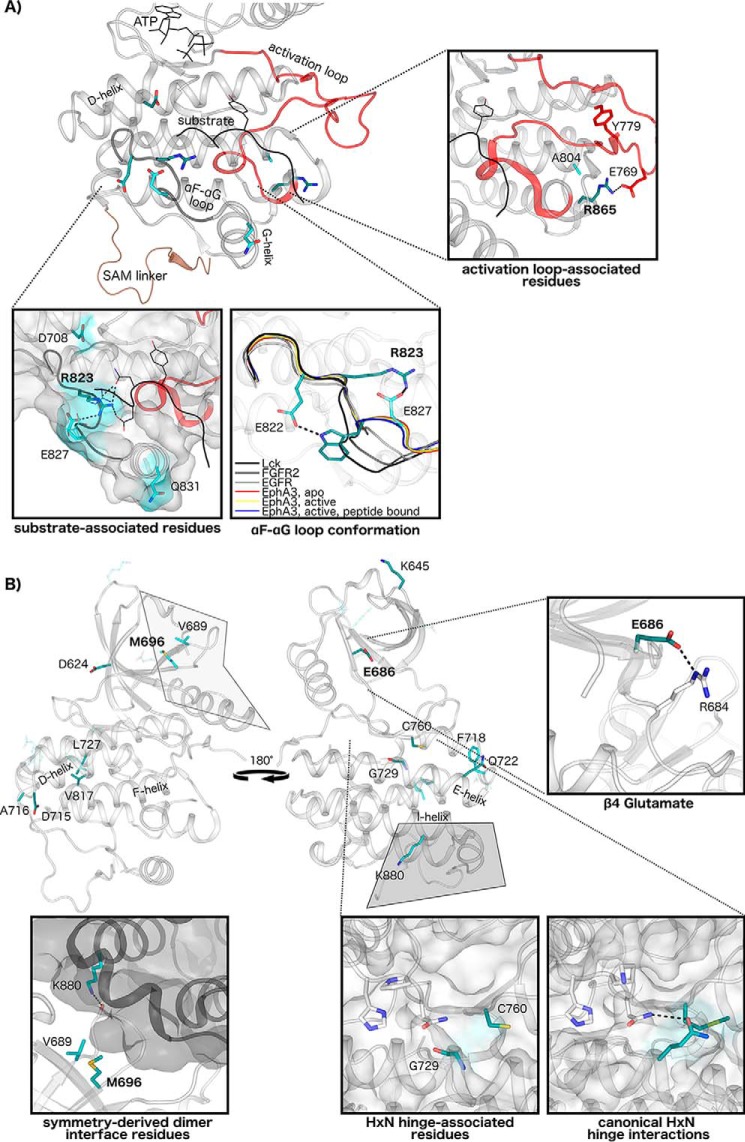
Structural interactions of Eph-specific residues of unknown function. A, Eph-specific residues contribute to unique interactions in the substrate-binding region and activation loop. Key conserved residues in this region are mapped to structures of human EphA3, with darker shading indicating greater sequence constraints at that position. ATP and peptide substrate are shown in black, the activation loop is in red, and the SAM domain linker is in brown. Interactions of Eph-specific residues and peptide substrate are shown, with different rotamer conformations between peptide-bound (shown as sticks) (Protein Data Bank code 3FXX) and peptide-unbound structures (shown as lines) (Protein Data Bank code 2QOC). The unique conformation of the αF-αG loop in Eph structures is shown relative to other tyrosine kinase structures: EphA3, apo (Protein Data Bank code 2QO2); EphA3, active (Protein Data Bank code 2QOC); EphA3, active, peptide-bound (Protein Data Bank code 3FXX), Lck (Protein Data Bank code 3LCK); FGFR2 (Protein Data Bank code 2PVF); and epidermal growth factor receptor (EGFR) (Protein Data Bank code 2GS2). Interactions of Eph-specific residues Arg865 and Ala804 with the activation loop are shown (EphA2, Protein Data Bank code 4TRL). B, Eph-specific residues in the N-lobe and hinge region. Interactions of the β4 glutamate are shown (Protein Data Bank code 2QOC). The N-lobe-to-C-lobe interface in a symmetry-derived dimer observed in several EphA3 structures is shown with key Eph-specific residues in the interface (Protein Data Bank code 2QOC) highlighted. Lys880 of this network forms a hydrogen bond to a backbone residue in the β4-β5 loop in the dimeric partner. Comparisons between canonical HXN hinge network interactions, which are conserved across diverse protein kinases, and the Eph family HXN hinge network are shown from representative structures (FGFR2, Protein Data Bank code 2PVF; EphA3, Protein Data Bank code 2QOC).