Abstract
Ethanol causes fetal alcohol spectrum disorders (FASDs) partly by inhibiting cell adhesion mediated by the L1 neural cell adhesion molecule. Ethanol interacts with an alcohol binding pocket in the L1 extracellular domain (ECD), and dephosphorylation of S1248 in the L1 cytoplasmic domain (CD) renders L1 adhesion insensitive to inhibition by ethanol (L1 insensitive). The mechanism underlying this inside-out signaling is unknown. Here we show that phosphorylation of the human L1-CD at S1152, Y1176, S1181, and S1248 renders L1 sensitive to ethanol by promoting L1 coupling with ankyrin-G and the spectrin-actin cytoskeleton. Knockdown of ankyrin-G or L1 mutations that uncouple L1 from ankyrin reduce L1 sensitivity to ethanol, but not methanol, consistent with a small conformational change in the extracellular alcohol binding pocket. Phosphorylation of Y1176 and ankyrin-G coupling with L1 are higher in NIH/3T3 clonal cell lines in which ethanol inhibits L1 adhesion than in ethanol-resistant NIH/3T3 clonal cell lines. Similarly, phosphorylation of Y1176 is higher in C57BL/6J mice that are sensitive to ethanol teratogenesis than in ethanol resistant C57BL/6N mice. Finally, polymorphisms in genes that encode ankyrin-G and p90rsk, a kinase that phosphorylates S1152, are linked to facial dysmorphology in children with heavy prenatal ethanol exposure. These findings indicate that genes that regulate L1 coupling to ankyrin may influence susceptibility to FASD.—Dou, X., Menkari, C., Mitsuyama, R., Foroud, T., Wetherill, L., Hammond, P., Suttie, M., Chen, X., Chen, S.-Y., Charness, M. E., Collaborative Initiative on Fetal Alcohol Spectrum Disorders. L1 coupling to ankyrin and the spectrin-actin cytoskeleton modulates ethanol inhibition of L1 adhesion and ethanol teratogenesis.
Keywords: FASD, alcohol, phosphorylation, susceptibility
Fetal alcohol spectrum disorder (FASD) is the most common preventable cause of intellectual disability (1, 2). At the most severe end of the spectrum are children with fetal alcohol syndrome (FAS), a disorder marked by typical facial dysmorphology, pre- and/or postnatal growth deficiency, neurobehavioral impairment, and structural or functional brain abnormalities. The facial dysmorphology of FAS arises from alcohol exposure during gastrulation, but ethanol can disrupt brain development throughout gestation (3). Consequently, many children with prenatal alcohol exposure and normal facial morphology still demonstrate cognitive, behavioral, and social impairment (2). Although FASD is preventable, fetal alcohol exposure still occurs frequently before pregnancy recognition and often later in gestation, despite the known risks (4).
Genetic factors modify the risk of FASD (5). Ethanol teratogenicity differs markedly in genetically related animal strains (6), and human monozygotic twins demonstrate higher concordance for FAS than dizygotic twins (7). The severity of FASD is a function of peak blood alcohol concentrations (8), and, not surprisingly, genetically regulated differences in the rate of alcohol metabolism influence the risk for FASD (5). Likewise, ethanol may unmask haploinsufficiency in developmentally important signaling pathways (9, 10). Ethanol greatly increases the rate of craniofacial abnormalities in zebrafish with haploinsufficiency of PDFGRA, and PDGFRA is linked to facial dysmorphology in children with heavy gestational alcohol exposure (10). These findings demonstrate important gene × alcohol interactions in the development of FASD, including the potential for ethanol to induce clinical expression of normally silent genotypes.
We have proposed a novel mechanism for gene × alcohol interactions: genetic regulation of the sensitivity to ethanol of a developmentally critical target protein—the L1 neural cell adhesion molecule (6). Ethanol may cause FASD partly by inhibiting L1-mediated cell adhesion (11) and neurite outgrowth (12). Ethanol inhibits L1 adhesion through interactions with a binding site in the L1 extracellular domain (ECD) (13), and ethanol inhibition of L1 adhesion is abolished by reducing phosphorylation of serine-1248, an ERK2 substrate in the L1 cytoplasmic domain (CD) (6). Of note, C57BL/6N (6N) mouse embryos have lower levels of ERK2 activity and are less susceptible to ethanol teratogenesis (ethanol-resistant embryos) than genetically related C57BL/6J (6J) mouse embryos (ethanol-sensitive embryos), suggesting that this inside-out signaling mechanism may partly mediate differences in the genetic susceptibility of these mouse substrains to FASD.
It is unknown how phosphorylation of the L1-CD alters ethanol interactions with a binding pocket far removed in the L1-ECD or whether genetically mediated differences in the activity of kinases that phosphorylate the L1-CD govern susceptibility to FASD in humans. Here we show that phosphorylation of 4 residues in the L1-CD regulates L1 sensitivity to ethanol by promoting L1 coupling to ankyrin-G and the spectrin-actin cytoskeleton. Loss of this cytoskeletal anchoring leads to the exclusion of ethanol, but not methanol, from an alcohol binding pocket in the L1-ECD. Phosphorylation of these obligate L1 intracellular phosphorylation sites (OLIPSs) and ankyrin-G association with L1 are correlated with L1 sensitivity to ethanol in clonal cell lines and in 6J and 6N mice. Finally, polymorphisms in the genes that encode ankyrin-G and one of the kinases that phosphorylates the OLIPSs are linked to facial dysmorphology in children with heavy ethanol exposure during pregnancy.
MATERIALS AND METHODS
Cell culture
We studied 2 NIH/3T3 clonal cells that stably express human L1 (hL1) in which L1-mediated cell adhesion (L1 adhesion) is sensitive (2A2-L1s, ethanol-sensitive clonal cell lines) or insensitive (3C3-L1i, ethanol-insensitive clonal cell lines) to ethanol (14). We also studied NIH/3T3 cells transiently transfected with hL1 constructs (15) bearing various L1 mutations (Supplemental Table 1). Cells were cultured in DMEM plus 10% bovine serum at 37°C with 10% CO2 atmosphere as described (6, 16). Transient transfection with plasmid DNA containing L1-wild type (WT) or mutant L1 was performed using PolyFect from Qiagen (Germantown, MD, USA) according to the manufacturer’s manual.
Reagents
The following antibodies were used: goat pAb SC-1508 (RRID AB_631086) against the L1-CD; mAb SC-53386 (UJ127, RRID AB_628937) against the L1-ECD (17), measuring L1 expression in whole cells; mAb SC-59868 (74-5H7, RRID AB_781976), measuring L1 dephospho-Y1176 (18); rabbit pAb SC-28561 (RRID AB_633909) against C terminus of ankyrin-G; mAb SC-46696 (RIID AB_671135) generated against a peptide derived from spectrin α II; goat pAb SC-1616 (RRID AB630836) against actin; and rabbit pAb SC-9104 (RRID AB_2241191) against β-tubulin (all from Santa Cruz Biotechnology, Santa Cruz, CA, USA); and rabbit pAb 4023S (RRID AB_10698604) for Fyn and mAb 2110S (RRID AB_10691385) for p60src (Cell Signaling Technology, Danvers, MA, USA). 5G3 mAb, which recognizes an epitope overlapping the Ig1 and Ig2 domains of L1 (17), was produced by Maine Biotechnology Services (Portland, ME, USA). Horseradish peroxidase–conjugated secondary antibodies against mouse (115-035-062, RRID AB_2338504), rabbit (711-035-152, RRID AB_10015282), and goat (705-035-003, RRID AB_2340390) were from Jackson ImmunoResearch Laboratories (West Grove, PA, USA).
Protein analysis
NIH/3T3 cells were grown to 75–85% confluence, harvested in PBS plus 2 mM EDTA, and pelleted by centrifugation (16). Cells were lysed in NP40 lysis buffer plus Halt Protease and Phosphatase Inhibitor Cocktail (1862495; Thermo Fisher Scientific, Waltham, MA, USA). The insoluble fraction was removed by centrifugation at maximum speed in an eppendorf centrifuge. Denatured proteins were separated and analyzed by Western blot. For L1 immunoprecipitation, whole cell lysates were incubated with mAb 5G3 (Maine Biotechnology Services, Portland, ME, USA) at 4°C for 2–4 h, and protein A–agarose beads were added to precipitate the antigen–antibody complex. Proteins were separated on SDS-PAGE and transferred to nitrocellulose membrane or PVDF for detection of protein using corresponding primary and secondary antibodies. Images of protein bands were acquired on an Amersham Imager 600 (GE Life Sciences, Pittsburgh, PA, USA), and densitometry was quantified using Image J (19). All data were normalized to 2A2-L1s cells or L1-WT transfected cells and plotted as means ± sem.
Short interfering RNA transfection of 2A2-L1s cells
Cells were transfected with short interfering RNAs (siRNAs) for Fyn (SC-35425), Src1 (SC-36556), ankyrin-G (SC-43268), and scrambled siRNA (SC-37007) as control (Santa Cruz Biotechnology) using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) following the manufacturers’ instructions (6). Forty-eight hours after transfection, cells were harvested for cell aggregation assays, Western blots, and flow cytometry (6). Levels of protein expression in Western blots were normalized to values from 2A2-L1s cells treated with scrambled siRNA.
Flow cytometry
Cell surface expression of L1 was quantified with flow cytometry using mAb 5G3 (6, 16). Data were presented as the geometric means ± sem.
Cell aggregation assay
2A2-L1s or 3C3-L1i cells stably expressing hL1 or NIH/3T3 cells transiently transfected with L1 constructs were harvested for cell aggregation assays, as described (6, 16). Cell aggregation assays were performed in the absence and presence of 100 mM ethanol or 100 mM methanol. L1 adhesion in NIH/3T3 cells was determined by separating cells into single-cell suspensions, agitating for 10 min at room temperature, and measuring the percentage of adherent cells using phase contrast microscopy, as described (16, 20). The half-maximal concentration of ethanol inhibition of L1 adhesion is 5 mM (16). Supramaximal concentrations of alcohols were used to identify the effects of mutations and inhibitors on alcohol inhibition. When comparing the effects of various treatments, alcohol inhibition of L1 adhesion was normalized to values obtained in 2A2-L1s cells. When evaluating the effect of various L1 mutations, alcohol inhibition of L1 adhesion in cells transiently transfected with mutant L1 constructs was normalized to values obtained in cells transfected with L1-WT.
Mouse embryo collection
C57BL/6J (6J) mice were obtained from The Jackson Laboratory (Bar Harbor, ME, USA) and C57BL/6N (6N) from Harlan Laboratories (Indianapolis, IN, USA). Pregnant mice were euthanized on gestational day (GD)10, and embryos of comparable developmental stage from several litters were collected for protein preparation as described (6). Studying GD 10 mouse embryos also allowed us to compare findings on Y1176 phosphorylation with our published data on MAPK activity in ethanol-sensitive and ethanol-resistant C57BL/6 mouse embryos (6). Western blot analysis was conducted on whole cell lysates using goat pAb SC-1508 to measure L1 expression and mAb 74-5H7 to measure L1-dephospho-Y1176. Goat pAb sc-1616 against actin was used as a loading control. Protein from 1 embryo was used for each lane and actin immunolabeling served as a within-lane control for each L1 antibody. Dephosphorylation of L1-Y1176 for each embryo was calculated as the ratio of 74-5H7 to UJ127 immunolabeling. These individual values were then normalized to the average level of dephosphorylation for all 6J and 6N embryos. All animal experiments were conducted with approval of the University of Illinois Institutional Animal Care and Use Committee.
Association analysis in the Collaborative Initiative on Fetal Alcohol Spectrum Disorders
Facial dysmorphology was characterized by dense surface modeling of 3-dimensional images (21). Principal components (PCs) were derived by reducing 28,000 data points using a mathematical representation resulting in 10 facial, 10 philtral, and 10 profile PCs. Statistical analyses were performed in PLINK (http://zzz.bwh.harvard.edu/plink/) and were limited to the 236 Collaborative Initiative on Fetal Alcohol Spectrum Disorders (CIFASD) participants delineated by single-nucleotide polymorphism (SNP) genotypic data to be of Caucasian ancestry. Genome-wide SNP array data were generated using Illumina arrays. Bonferroni correction for testing 30 PCs and 7 genes resulted in a corrected α = 2.4 × 10−4.
Statistical analysis
Data are expressed as means ± sem, except as noted. Statistical analyses were performed with ANOVA, and significant results (P < 0.05) were followed up using a 2-tailed Student’s t test for pairwise comparisons of interest. All analyses used Prism (GraphPad Software, La Jolla, CA, USA).
RESULTS
Phosphorylation of Y1176 is required for ethanol inhibition of L1 adhesion
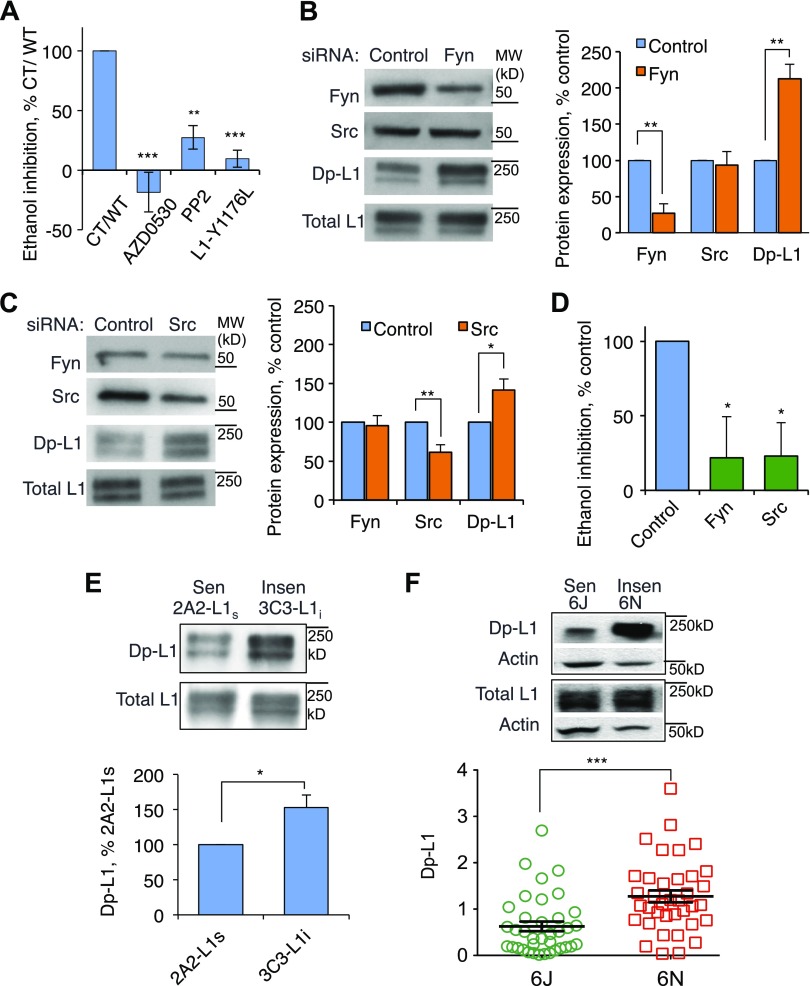
The observation that a reduction in ERK2 phosphorylation of S1248 abolishes L1 sensitivity to ethanol prompted a search for other OLIPSs. The Src family kinase (SFK) p60src phosphorylates L1 at Y1176 (18) and is necessary for L1-mediated neurite outgrowth in cerebellar granular neurones (CGNs) (22). To determine whether Y1176 is an OLIPS, we inhibited SFK with AZD0530 or PP2 in 2A2-L1s cells, a clonal NIH/3T3 cell line stably transfected with hL1. Treatment with 10 nM AZD0530 or 20 µM PP2 reduced Y1176 phosphorylation and L1 sensitivity to ethanol without affecting the expression, subcellular distribution, or adhesion of L1 (Fig. 1A; Supplemental Fig. 1). To verify that Y1176 is an OLIPS, we transiently transfected NIH/3T3 cells with an hL1 construct in which Y1176 was replaced with leucine, a residue that cannot be phosphorylated. The L1-Y1176L construct showed normal expression and adhesion but was insensitive to ethanol (Figs. 1A; Supplemental Fig. 1C).
Figure 1.
SFK activity and phosphorylation of L1-Y1176 regulate L1 sensitivity to ethanol. A) 2A2-L1s cells stably expressing hL1 were incubated for 1 h in DMEM supplemented with the SFK inhibitors 10 nM AD0530 or 20 μM PP2 and harvested for cell aggregation assays. NIH/3T3 cells were transiently transfected with hL1 in which Y1176 was replaced with leucine. Cells were harvested for cell aggregation assays performed in the absence and presence of 100 mM ethanol, which is a supramaximally effective concentration. Ethanol inhibition of L1 adhesion was normalized to values obtained in 2A2-L1s cells unexposed to SFK inhibitors (CT) or cells transfected with L1-WT. Shown are means ± sem levels of ethanol inhibition from 4 to 17 experiments (F = 14.54; P = 0.0004). **P < 0.01; ***P < 0.001, compared with CT or WT. B, C) Fyn-siRNA (B) and Src-siRNA (C) specifically reduced phosphorylation of Y1176 in 2A2-L1s cells. Protein expression was quantified using Western blots and the following antibodies against Src and Fyn: antibody UJ127, which recognizes total L1, and mAb 74-5H7, which recognizes dephospho-L1-Y1176 (Dp-L1). For quantitative densitometry of protein expression, values for Src, Fyn, and dp-L1 expression were normalized to values for total L1 obtained from the same gel lanes. Expression of proteins in the presence of specific siRNAs was then normalized to expression of the same proteins in the presence of scrambled siRNA. Shown are means ± sem levels of the indicated proteins from 4 to 8 independent experiments (F = 21.26; P < 0.0001). *P < 0.05; **P < 0.01. D) Data from cells treated with Fyn or Src siRNA were normalized to values from cells treated with scrambled siRNA (control). Shown are means ± sem levels of ethanol inhibition from 8 independent experiments (F = 6.39; P = 0.0068). *P < 0.025 compared with control. E) Levels of L1-dephospho-Y1176 in ethanol-sensitive 2A2-L1s and ethanol-insensitive 3C3-L1i cells. Upper panel shows a representative Western blot using mAb 74-5H7 and goat pAb SC-1508, which recognizes total L1. Lower panel shows mean ± sem levels of dephospho-Y1176 in 3C3-L1i cells normalized to values from 2A2-L1s cells from 10 independent experiments (t = 3.03). *P < 0.025. F) Levels of dephospho-Y1176 in GD-10 embryos from ethanol-sensitive C57BL/6J (6J) and ethanol-resistant C57BL/6N (6N) mice. Upper panel shows a representative Western blot using mAb 74-5H7 from 1 6J and 1 6N embryo. Lower panel shows individual and mean ± sem levels of L1-dephospho-Y1176 in 6N compared with 6J embryos. For each embryo, levels of L1-dephospho-Y1176 and total L1 were separately normalized to levels of actin within the same lane. These individual values were then normalized to the average level of dephosphorylation for all 6J and 6N embryos (t = 3.91). ***P < 0.001 (n = 37).
Src and Fyn contribute to L1-Y1176 phosphorylation
Because Fyn is abundantly expressed in NIH/3T3 cells and in CGNs (23), we asked whether Fyn also affects the phosphorylation of Y1176. 2A2-L1s cells were transfected with a Fyn or Src siRNA to selectively decrease the expression of Fyn or Src. siRNA knockdown of SRC decreased p60src expression by ∼40% without reducing Fyn (Fig. 1B). Similarly, siRNA knockdown of FYN decreased Fyn expression more than 3-fold without reducing p60src expression (Fig. 1C). Neither siRNA affected L1 expression or L1 adhesion (Fig. 1B, C; Supplemental Fig. 2), but each decreased Y1176 phosphorylation and ethanol inhibition of L1 adhesion (Fig. 1D). Taken together, these findings indicate that phosphorylation of Y1176 by p60src or Fyn is necessary for ethanol inhibition of L1 adhesion.
Differential ethanol sensitivity of hL1-expressing clonal NIH/3T3 cell lines and mouse substrains is associated with differential phosphorylation of Y1176
We showed previously that stable transfection of NIH/3T3 cells with hL1 yields clonal cell lines that are either sensitive (2A2-L1s) or insensitive (3C3-L1i) to ethanol (14). Because phosphorylation of Y1176 is necessary for L1 sensitivity to ethanol, we asked whether levels of Y1176 phosphorylation differed in these 2 clonal cell lines. Indeed, phosphorylation of Y1176 was significantly higher in ethanol-sensitive 2A2-L1s cells than in ethanol-insensitive 3C3-L1i cells (Fig. 1E). We also measured phosphorylation of Y1176 in 6J and 6N mouse embryos. The average level of Y1176 phosphorylation in GD10 whole embryos was significantly higher in the ethanol-sensitive 6J mice than in the ethanol-resistant 6N mice (Fig. 1F). These data demonstrate that increased phosphorylation of Y1176 is associated with increased ethanol inhibition of L1 adhesion in clonal NIH/3T3 cell lines and greater sensitivity to ethanol teratogenesis in genetically related C57BL/6 mouse substrains.
L1 sensitivity to ethanol requires phosphorylation at 4 separate residues in the L1-CD
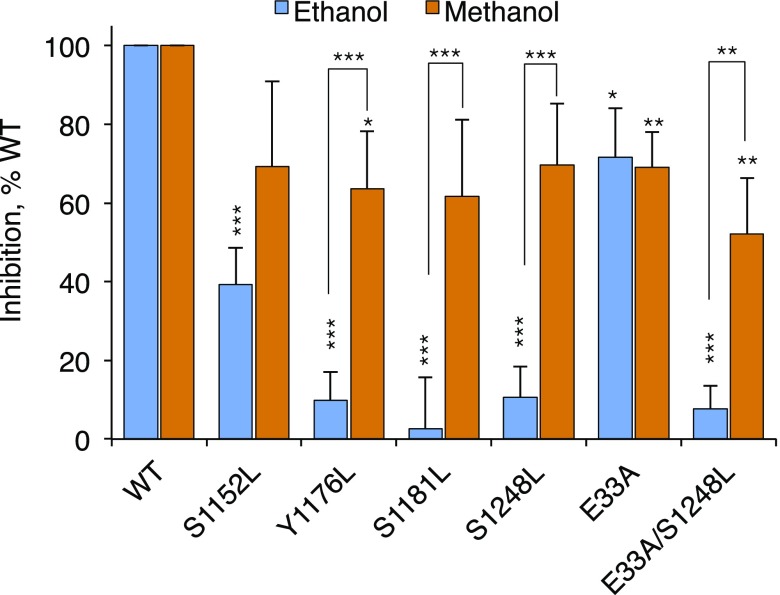
To identify other OLIPS, we studied constructs in which leucine replaced S1152, a substrate for p90rsk (24), or S1181, a substrate for casein kinase II (25). Neither mutation reduced the expression, subcellular distribution, or adhesion of L1 (Supplemental Fig. 3). Ethanol inhibition of L1 adhesion was significantly reduced by the S1152L mutation and was abolished by the S1181L mutation (Fig. 2). Hence, L1 sensitivity to ethanol requires phosphorylation of at least 4 OLIPS (S1152, Y1176, S1181, and S1248).
Figure 2.
Inhibition of L1 inhibition by ethanol and methanol in L1 constructs bearing leucine mutations of 4 OLIPS. NIH/3T3 cells were transiently transfected with hL1 in which residues were replaced with leucine or alanine, as indicated. Cells were harvested for cell aggregation assays performed in the absence and presence of 100 mM ethanol or 100 mM methanol. Alcohol inhibition of adhesion in mutant constructs was normalized to values obtained in L1-WT. Shown are means ± sem levels of alcohol inhibition from 9 to 142 independent experiments (F = 9.87; P < 0.0001). *P < 0.05; **P < 0.01; ***P < 0.001 comparing inhibition in mutants with WT or comparing ethanol with methanol (brackets).
L1 sensitivity to methanol is preserved in leucine mutations of the OLIPS
1-Alcohols of up to 4 carbons (1-butanol) inhibit L1 adhesion, whereas 1-alcohols of >4 carbons have no effect (11). This alcohol cutoff indicates a size limitation for alcohol interactions with L1. We asked whether the elimination of phosphorylation at the OLIPS reduced sensitivity not just for ethanol but also for the smaller alcohol methanol. Interestingly, methanol inhibition of L1 adhesion was preserved after leucine replacement of each OLIPS (Fig. 2). These findings suggest that dephosphorylation of each OLIPS narrows, but does not occlude, an alcohol binding pocket within the L1-ECD.
Photolabeling experiments with azibutanol identified an alcohol binding pocket bounded by 2 closely approximated alcohol binding residues on the L1-ECD: E33 on Ig1 and Y418 on Ig4 (13). Alanine mutation of E33 (E33A) increased the 1-alcohol cutoff from 4 carbons to 10 (16), but adding the S1248L mutation (1-carbon cutoff) to a construct bearing the E33A mutation (10-carbon cutoff) yielded an alcohol cutoff of just 1 carbon (Fig. 2), suggesting that this L1-CD mutation altered the conformation of the previously identified alcohol binding pocket in the L1-ECD.
Association of L1 with ankyrin-G and the spectrin-actin cytoskeleton is necessary for L1 sensitivity to ethanol
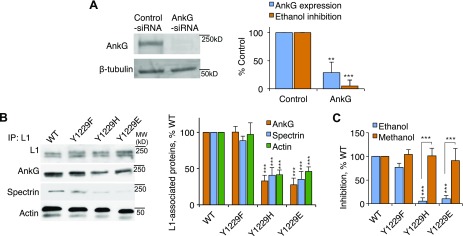
One potential molecular transducer for this inside-out signaling is ankyrin. Ankyrin binds reversibly to the L1 family of cell adhesion molecules at a highly conserved consensus sequence (FIGQY), thereby linking L1 to the spectrin-actin cytoskeleton (26, 27). Knockdown of ankyrin-G using siRNA did not reduce L1 adhesion but eliminated ethanol inhibition of L1 adhesion (Fig. 3A; Supplemental Fig. 4). Knockdown of ankyrin-B reduced L1 adhesion, precluding further testing of ethanol inhibition of L1 adhesion (data not shown). These findings indicate that ankyrin-G expression is required for L1 sensitivity to ethanol.
Figure 3.
Effect of L1 association with ankyrin-G on alcohol inhibition of L1 adhesion. A) Effect of ankyrin-G (AnkG) and control (scrambled) siRNA on the expression of ankyrin-G and ethanol inhibition of L1 adhesion in 2A2-L1s cells. Upper panel: Representative Western blot showing immunolabeling of ankyrin-G and β-tubulin from whole cell lysates. Lower panel: Densitometry quantification of ankyrin-G expression (light bars). The expression of ankyrin-G was normalized to β-tubulin. Ankyrin-G expression and ethanol inhibition of L1 adhesion (dark bars) in cells treated with ankyrin-G siRNA were normalized to values obtained in cells treated with control siRNA. Shown are means ± sem levels of ankyrin-G expression (t = 6.71; **P < 0.01) and ethanol inhibition (t = 7.33, ***P < 0.001) from 6 to 17 independent experiments. B) Effect of mutations of L1-Y1229 on L1 association with ankyrin-G, spectrin, and actin in NIH/3T3 cells. Left panel: Representative gel. Right panel: Densitometry analysis of L1 association with each protein. Densities of bands for ankyrin-G, spectrin, and actin were normalized to values for total L1, and values in cells expressing mutant proteins were expressed as a percentage of values in cells expressing L1-WT. Shown are mean percentages ± sem from 5 to 8 independent experiments (F = 15.98; P < 0.0001). ***P < 0.001. C) Effect of Y1229 mutation on alcohol inhibition of L1 adhesion. Shown is the mean ± sem inhibition of L1 adhesion by ethanol and methanol for L1-Y1229 mutant–expressing cells in relation to L1-WT–expressing cells from 8 to 142 independent experiments (F = 16.09; P < 0.0001). ***P < 0.001.
Ankyrin binding to L1 requires dephosphorylation of FIGQY1229 (28, 29). To determine whether ankyrin-G binding to L1 is necessary for L1 sensitivity to ethanol, we used immunoprecipitation to study ankyrin association with 3 previously described L1 mutations (30, 31). L1-Y1229F, which cannot be phosphorylated at the 1229 position, associated normally with ankyrin-G, spectrin, and actin (Fig. 3B). In contrast, L1-Y1229H, a pathogenic human mutation (32) that also cannot be phosphorylated at the 1229 position, associated poorly with ankyrin-G, spectrin, and actin. Finally, the Y1229E mutation, which mimics permanent phosphorylation at the 1229 position, associated poorly with ankyrin-G, spectrin, and actin. None of these mutations changed the expression or adhesion of L1 (Supplemental Fig. 3). Ethanol inhibited L1 adhesion in L1-Y1229F–transfected cells but had no effect on the adhesion of cells transfected with L1-Y1229H or L1-Y1229E (Fig. 3C). In contrast, methanol inhibited the adhesion of both L1-Y1229H and L1-Y1229E. These findings indicate that association of L1 with ankyrin-G and the spectrin-actin cytoskeleton is necessary for ethanol inhibition of L1 adhesion. Of note, mutations that reduce ankyrin-G association with L1 produced the same shift in the alcohol cutoff as leucine mutation of each of the 4 OLIPS.
OLIPS phosphorylation promotes L1 association with ankyrin-G and the spectrin-actin cytoskeleton
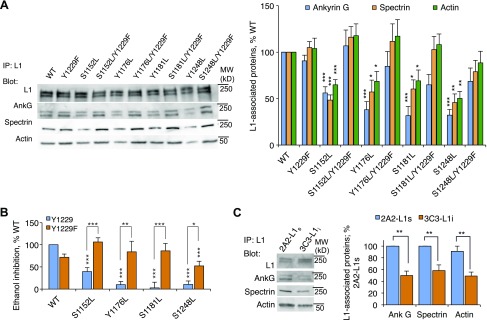
We next asked whether dephosphorylation of OLIPS blocks L1 sensitivity to ethanol by decreasing ankyrin-G binding to L1 and L1 association with the spectrin-actin cytoskeleton. Figure 4A shows that the S1152L, S1181L, Y1176L, and S1248L mutations each significantly reduced L1 association with ankyrin-G, spectrin, and actin. Furthermore, L1 association with the spectrin-actin cytoskeleton and L1 sensitivity to ethanol could be restored in these constructs by adding a second mutation, Y1229F, which promotes ankyrin-G binding to L1 (Fig. 4A, B). These results imply that OLIPS phosphorylation promotes L1 sensitivity to ethanol by inducing the association of L1 with ankyrin-G and the spectrin-actin cytoskeleton.
Figure 4.
Effect of OLIPS mutations on L1 association with ankyrin and L1 sensitivity to ethanol. A). Representative Western blot and densitometric analysis of L1 association with ankyrin-G, spectrin, and actin in cells expressing single OLIPS leucine mutations in the absence and presence of the Y1229F mutation (F = 7.72; P < 0.0001). *P < 0.05; **P < 0.01; ***P < 0.001 (n = 6–17). B) Ethanol inhibition of L1 adhesion in the constructs shown in A (F = 9.27; P < 0.0001). *P < 0.05; **P < 0.01; ***P < 0.001 (n = 9–142). C) Association of ankyrin-G, spectrin, and actin with L1 in 2A2-L1s cells vs. 3C3-L1i cells (F = 10.69; P < 0.0001). **P < 0.01 (n = 6–11).
L1 is differentially associated with the spectrin-actin cytoskeleton in ethanol-sensitive and ethanol-insensitive clonal cell lines
We showed previously that MAPK activity and L1-Y1176 phosphorylation were higher in ethanol-sensitive 2A2-L1s cells than in ethanol-insensitive 3C3-L1i cells (6). Figure 4C demonstrates that L1 association with ankyrin-G, spectrin, and actin was also significantly higher in 2A2-L1s cells than in 3C3-L1i cells. These findings provide additional evidence that OLIPS phosphorylation influences L1 sensitivity to ethanol through downstream actions that promote ankyrin-G binding to L1 and L1 anchoring to the spectrin-actin cytoskeleton.
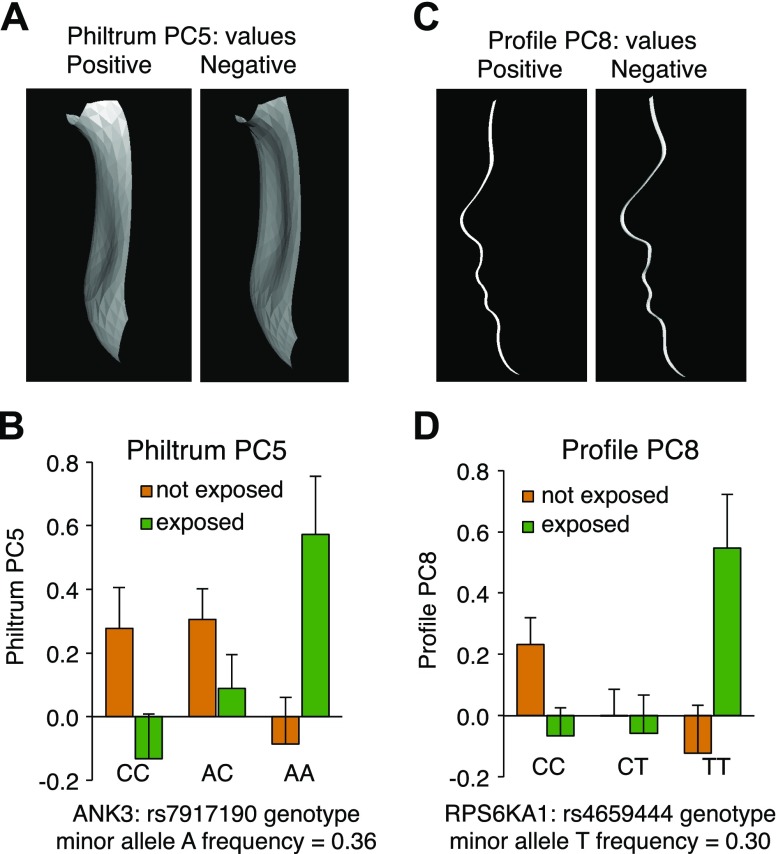
Variation in ANK3 and RPS6KA1 is associated with features of FASD
If L1 sensitivity to ethanol is modulated by OLIPS phosphorylation and the reversible association of L1 with ankyrin-G, then genetic variation in the pathways that mediate these events might influence genetic susceptibility to FASD in humans. We used genome-wide association study data generated as part of the assessment of participants in the CIFASD (10) and limited our exploration to association of SNPs in MAPK1 (ERK2), SRC (p60Src), FYN (Fyn kinase), RPS6KA1 (p90rsk), CSNK2A1 (CK2), ANK3 (ankyrin-G), and EPHB2 (EphB2) on facial phenotypes. Statistical analyses tested for the effect of a SNP × alcohol exposure interaction on a series of facial, philtral, and profile phenotypes. Evidence suggesting an SNP × alcohol exposure interaction was observed for 2 of the 7 genes (ANK3 and RPS6KA1) (Fig. 5). There was no evidence of an SNP × alcohol exposure interaction for SRC, FYN, EPHB2, MAPK1, or CSNK2A1.
Figure 5.
SNP × alcohol interaction in 2 genes that regulate L1 sensitivity to ethanol. A) Philtrum PC5 phenotypic extremes. Positive philtrum values are consistent with features associated with FASD. B) ANK3 and philtrum PC5 interaction. Individuals with the AA genotype at rs7917190 who were exposed to alcohol prenatally had a significantly flatter upper philtrum compared with AA individuals not exposed to alcohol who had a groove in the philtrum [p (SNP × alcohol exposure) = 3.9 × 10−4]. C) Profile PC8 phenotypic extremes. Positive profile values are consistent with features associated with FASD. D) RPS6KA1 and profile PC8 interaction. Individuals with the TT genotype at rs4659444 who were exposed to alcohol prenatally had significantly greater flattening of the midfacial profile and micrognathia compared with TT individuals not exposed to alcohol who had a longer and more pronounced nasal profile and no micrognathia [p (snp × alcohol exposure) = 2.2 × 10−5].
DISCUSSION
Ethanol disruption of L1 function has been implicated in the pathogenesis of FASD. FAS is a partial phenocopy of human disorders caused by mutations in the L1 gene, with significant overlap in neuropathology (11, 33). Several pathogenic human L1 gene mutations disrupt L1 homophilic adhesion or L1-mediated neurite outgrowth (34, 35). Likewise, concentrations of ethanol attained in the fetus after maternal ingestion of 1 or 2 drinks also disrupt L1-mediated cell–cell adhesion in neural cell lines, transfected fibroblast cell lines, and cerebellar granule neurons (11) and inhibit L1-mediated neurite outgrowth in cerebellar granule neurons (12). The connection between L1 and FASD is further supported by the finding that peptides and small molecules that block ethanol inhibition of L1 adhesion prevent ethanol-induced apoptosis, growth retardation, and neural tube defects in mouse embryos (36–38).
Early research on alcohol and L1 disclosed that not all L1-expressing cells are sensitive to ethanol (14, 39). A single transfection of NIH/3T3 cells with hL1 yielded clonal cell lines in which ethanol either inhibited (2A2-L1s) or did not inhibit (3C3-L1i) L1 adhesion (14). One factor that proved necessary for L1 sensitivity to ethanol was ERK2 phosphorylation of S1248 in the L1-CD, and overexpression of MEK1, the kinase that activates ERK2, rendered 3C3-L1i cells sensitive to ethanol (6). Our search for other OLIPS within the highly conserved L1-CD identified 3 other sites (S1152, Y1176, and S1181). Our search was targeted, and other OLIPS may exist.
Leucine mutation at each OLIPS reduced inhibition of L1 adhesion by ethanol, but not methanol, without altering the expression, subcellular distribution, or adhesion of L1. The same phenotype resulted from L1-Y1229 mutations that promote the dissociation of L1 from ankyrin. These findings suggested that OLIPS phosphorylation might influence L1 sensitivity to ethanol by promoting the reversible association of L1 and ankyrin. In support of this hypothesis, leucine mutation of each OLIPS reduced L1 association with ankyrin-G, spectrin, and actin. Furthermore, pairing each OLIPS leucine mutation with Y1229F, a mutation that promotes ankyrin binding to L1, restored L1 association with ankyrin-G, spectrin, and actin and rendered these double mutants sensitive to ethanol. These findings establish that OLIPS phosphorylation mediates L1 sensitivity to ethanol by promoting the association of L1 with ankyrin and the spectrin-actin cytoskeleton.
Ankyrin coupling with L1 requires dephosphorylation of Y1229 (27, 40); hence, OLIPS phosphorylation might inhibit the kinases or activate the phosphatases that act at this site. Consistent with this hypothesis, replacement of Y1229 with glutamic acid to mimic permanent phosphorylation did uncouple L1 from ankyrin-G, spectrin, and actin and rendered L1 insensitive to ethanol. EphB2 phosphorylates Y1229, but it does so by recruiting p60src and without a requirement for p60src phosphorylation of the OLIPS Y1176 (40). Conceivably, the association of p60src with Y1176 removes it from a microdomain where it can phosphorylate Y1229. We have shown previously that ethanol does not change the expression of p60src or its translocation into lipid rafts in NIH/3T3 cells (41).
Our data make clear that dephosphorylation of Y1229 is not a sufficient condition for L1 sensitivity to ethanol; the pathogenic mutation L1-Y1229H cannot be phosphorylated yet was insensitive to ethanol. This mutation also reduced L1 binding to ankyrin-G, presumably due to steric hindrance from the histidine residue. These findings underscore the importance of L1-ankyrin association as a critical determinant of L1 sensitivity to ethanol. Conditions that favor ankyrin-L1 association and render L1 adhesion sensitive to ethanol inhibition may also favor ethanol disruption of brain development. However, it is important to note that L1-ankyrin association is required for normal development (42) and promotes abnormal brain development only in the presence of ethanol.
How might the association of L1 with ankyrin-G mediate inside-out signaling? The anchoring of L1 by ankyrin to the spectrin-actin cytoskeleton restricts horizontal diffusion of L1 within the cell membrane and modifies L1 homophilic interactions (27, 28). These reversible changes in extracellular interactions provide evidence for reversible changes in the conformation of the L1-ECD. Our data do not rule out the possibility that OLIPS phosphorylation subtly alters the alcohol binding site by disrupting the homomeric clustering of L1 molecules in the plane of the membrane (17, 43). Inside-out signaling has been reported for phosphorylation of L1-T1172, which unmasks epitopes in the L1-ECD, modifies interactions of L1 with integrins, and alters L1-mediated migration of pancreatic cancer cells (17). It is unknown whether this mutation also affects L1 binding to ankyrin or L1 sensitivity to ethanol.
L1 homophilic adhesion is mediated by interactions between the extracellular domains of adjacent L1 molecules and is optimized when the Ig1 domain folds over the Ig4 domain to assume a horseshoe configuration (44). Homology modeling suggests that the alcohol binding residues E33 on Ig1 and Y418 on Ig4 are separated by just 2.8 Å at a location important for maintaining this horseshoe configuration (13). Mutation of these residues modifies the pharmacology of alcohol inhibition of L1 adhesion (16), providing further evidence that they contribute to a pharmacologically relevant alcohol binding pocket. Leucine OLIPS mutations and the histidine and glutamic acid mutations of Y1229 uncouple L1 from the spectrin-actin cytoskeleton, producing a shift in the alcohol cutoff from 4 carbons to 1. This shift implies a reduction in the size of the alcohol binding pocket by approximately the volume of 3 methyl groups. Importantly, when combined with the OLIPS mutation S1248L, the alcohol cutoff of the E33A mutation is reduced from 10 carbons to 1 (16). Taken together, these findings provide evidence that dissociation of L1 from the spectrin-actin cytoskeleton induces a small change in the conformation of the L1-ECD; this change is insufficient to modify L1 adhesion but is sufficient to exclude ethanol (but not methanol) from the alcohol binding pocket at the interface of Ig1 and Ig4 (Fig. 6). Hence, ethanol inhibits L1 adhesion and disrupts brain development by interacting with the L1-ECD only when L1 binds ankyrin in the L1-CD. This hypothesis is consistent with data from humans showing that point mutations in the L1-ECD that disrupt L1 adhesion may produce severe brain dysmorphology (45).
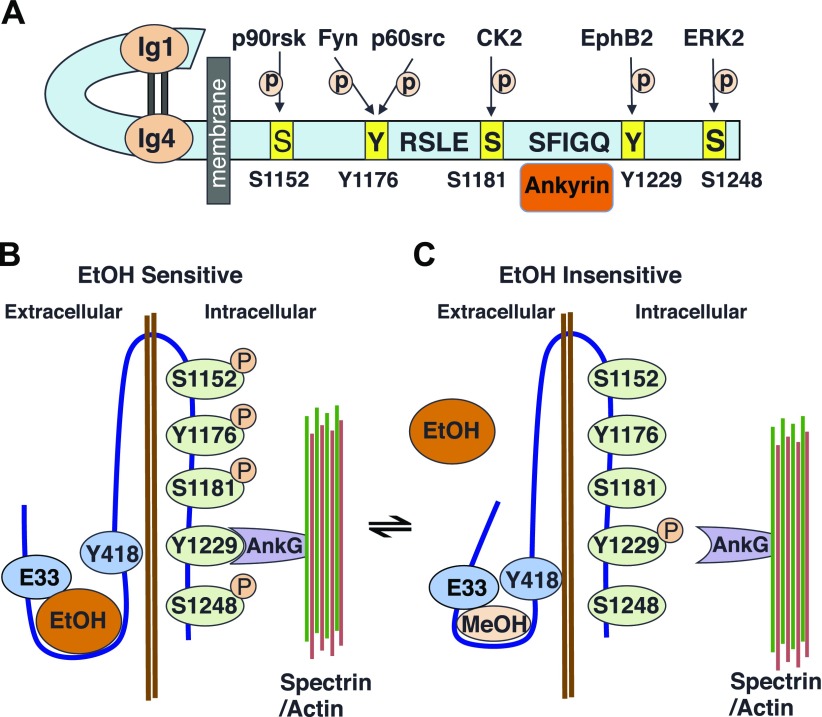
Figure 6.
Depiction of ankyrin interactions with L1 and their effect on ethanol inhibition of L1 adhesion. A) Schematic representation of the L1 molecule showing the 4 OLIPS, their associated kinases, and the ankyrin binding motif. B) Ethanol-sensitive state of L1. Phosphorylation of the 4 OLIPS coupled with dephosphorylation of Y1229 promotes the association of L1 with ankyrin and the spectrin-actin cytoskeleton and the interaction of ethanol with an alcohol binding pocket in the L1-ECD. C) Dephosphorylation of any of the OLIPs or phosphorylation of Y1229 leads to uncoupling of L1 from ankyrin and the spectrin-actin cytoskeleton and a small conformational change in the alcohol binding pocket, reducing access to ethanol but not methanol.
Our observation that loss of ankyrin-G binding to L1 does not affect L1 adhesion in NIH/3T3 cells contrasts with findings in neuroblastoma cells (28), where loss of ankyrin binding decreases cell aggregation mediated by neurofascin, a member of the L1 family of cell adhesion molecules (CAMs) (46). We did observe a reduction in L1 adhesion with siRNA knockdown of ankyrin-B. Hence, the magnitude of change in the conformation of the L1-ECD induced by ankyrin binding to L1 family CAMs appears to vary as a function of ankyrin isoform, cell type, and L1CAM family member.
L1 interactions with ankyrin-G may partly determine heritable differences in susceptibility to FASD. MAP kinase activity and phosphorylation of L1-Y1776 were greater in ethanol-sensitive 2A2-L1s cells and GD10 6J mouse embryos than in their ethanol-resistant counterparts (6). Both L1 and ankyrin-G are expressed at this early stage of development (36, 46). Likewise, association of L1 with ankyrin-G, spectrin, and actin is greater in 2A2-L1s cells than in 3C3-L1i cells. Both the clonal cell lines and the C57BL/6 mice are derived from common progenitor stock and likely underwent minor genetic drift through clonal selection and inbreeding, respectively. In this regard, they are valuable model systems for studying the effect of genetic variation on the response to prenatal ethanol exposure and for identifying candidate genes that determine susceptibility to FASD. Further work is required to determine whether the mechanisms underlying L1 sensitivity to ethanol in these model systems also mediate the differential sensitivity to ethanol of diverse neuronal cellular populations (47).
We studied the well-characterized CIFASD human cohort to learn whether genes that regulate the phosphorylation of OLIPS or L1 association with ankyrin-G are susceptibility genes for FASD. Because our cohort was relatively small, we evaluated just 7 candidate genes to minimize confounds from multiple comparisons. Polymorphisms in 2 of these 7 genes (RPS6KA1 and ANK3) demonstrated an association with facial dysmorphology in children who were heavily exposed to ethanol. These results are consistent with the hypothesis that pathways that regulate L1 sensitivity to ethanol also determine genetic susceptibility to prenatal ethanol exposure in humans. Alternatively, ethanol may be unmasking haploinsufficiency in 2 genes that regulate development through mechanisms that are unrelated or only partially related to L1, a phenomenon that has been demonstrated for other genes in zebrafish, mice, and humans (9, 10).
ANK3 is expressed as early as the 2-cell stage of development and plays an important role in cellular polarity (42, 46). Ankyrin-G expression continues throughout development and is required for the recruitment of voltage-gated sodium channels to the axon initial segment and the development of nodes of Ranvier (42). Mutations in ANK3 have been described in a small number of patients with autism, intellectual disability, sleep disorder, and attention deficit hyperactivity disorder (48, 49). Polymorphisms of ANK3 have also been associated with bipolar disorder, schizophrenia, and autism (50, 51). RPS6KA1 is highly expressed in the neural epithelium during early development and persists in the CGN layer in the adult brain, although its role in brain development is not well understood (52). Further work is required to determine whether and how genetic regulation of L1 coupling to ankyrin modifies FASD susceptibility in humans.
Supplementary Material
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
ACKNOWLEDGMENTS
The authors thank Valerie Novakovic and Jerry Lee (Veterans Affairs Boston Healthcare System; Harvard Medical School) for assistance with FACS measurements and critical review of the manuscript. Part of this work was done in conjunction with the Collaborative Initiative on Fetal Alcohol Spectrum Disorders (CIFASD), which is funded by grants from the U.S. National Institutes of Health, National Institute on Alcohol Abuse and Alcoholism (NIAAA). Additional information about CIFASD can be found at www.cifasd.org. This work was supported by NIAAA Grant R01AA012974 (to M.E.C.), NIAAA U24AA014811 as a component of CIFASD (to M.E.C.); The Medical Research Service and Veterans Affairs Merit Review 5I01BX002374 (to M.E.C.); Department of Defense Grant W81XWH-12-2-0048 Subaward 8742sc (to M.E.C.); NIAAA U01AA014809 (to T.F. and P.H.); NIAAA R01AA020265 (to S.-Y.C), NIAAA R01AA021434 (to S.-Y.C) and NIAAA P50AA024337 (to S.-Y.C). The authors declare no conflicts of interest.
Glossary
- CAM
cell adhesion molecule
- CD
cytoplasmic domain
- CGN
cerebellar granular neurone
- CIFASD
Collaborative Initiative on Fetal Alcohol Spectrum Disorders
- ECD
extracellular domain
- FASD
fetal alcohol spectrum disorder
- FAS
fetal alcohol syndrome
- GD
gestational day
- hL1
human L1
- OLIPS
obligate L1 intracellular phosphorylation site
- PC
principal component
- SFK
Src family kinase
- siRNA
short interfering RNA
- SNP
single-nucleotide polymorphism
- WT
wild type
Footnotes
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
AUTHOR CONTRIBUTIONS
X. Dou, T. Foroud, and M. Charness designed the research; X. Dou. C. Menkari, R. Mitsuyama, L. Wetherill, M. Suttie and X. Chen performed the research; X. Dou, T. Foroud, P. Hammond, S. Chen and M. Charness analyzed data, X. Dou, T. Foroud, P. Hammond, S. Chen and M. Charness wrote the paper.
REFERENCES
- 1.May P. A., Baete A., Russo J., Elliott A. J., Blankenship J., Kalberg W. O., Buckley D., Brooks M., Hasken J., Abdul-Rahman O., Adam M. P., Robinson L. K., Manning M., Hoyme H. E. (2014) Prevalence and characteristics of fetal alcohol spectrum disorders. Pediatrics 134, 855–866https://doi.org/10.1542/peds.2013-3319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoyme H. E., Kalberg W. O., Elliott A. J., Blankenship J., Buckley D., Marais A. S., Manning M. A., Robinson L. K., Adam M. P., Abdul-Rahman O., Jewett T., Coles C. D., Chambers C., Jones K. L., Adnams C. M., Shah P. E., Riley E. P., Charness M. E., Warren K. R., May P. A. (2016) Updated clinical guidelines for diagnosing fetal alcohol spectrum disorders. Pediatrics 138https://doi.org/10.1542/peds.2015-4256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lipinski R. J., Hammond P., O’Leary-Moore S. K., Ament J. J., Pecevich S. J., Jiang Y., Budin F., Parnell S. E., Suttie M., Godin E. A., Everson J. L., Dehart D. B., Oguz I., Holloway H. T., Styner M. A., Johnson G. A., Sulik K. K. (2012) Ethanol-induced face-brain dysmorphology patterns are correlative and exposure-stage dependent. PLoS One 7, e43067.https://doi.org/10.1371/journal.pone.0043067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Floyd R. L., Decouflé P., Hungerford D. W. (1999) Alcohol use prior to pregnancy recognition. Am. J. Prev. Med. 17, 101–107https://doi.org/10.1016/S0749-3797(99)00059-8 [DOI] [PubMed] [Google Scholar]
- 5.Eberhart J. K., Parnell S. E. (2016) The genetics of fetal alcohol spectrum disorders. Alcohol. Clin. Exp. Res. 40, 1154–1165https://doi.org/10.1111/acer.13066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dou X., Wilkemeyer M. F., Menkari C. E., Parnell S. E., Sulik K. K., Charness M. E. (2013) Mitogen-activated protein kinase modulates ethanol inhibition of cell adhesion mediated by the L1 neural cell adhesion molecule. Proc. Natl. Acad. Sci. USA 110, 5683–5688https://doi.org/10.1073/pnas.1221386110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Streissguth A. P., Dehaene P. (1993) Fetal alcohol syndrome in twins of alcoholic mothers: concordance of diagnosis and IQ. Am. J. Med. Genet. 47, 857–861https://doi.org/10.1002/ajmg.1320470612 [DOI] [PubMed] [Google Scholar]
- 8.Bonthius D. J., Goodlett C. R., West J. R. (1988) Blood alcohol concentration and severity of microencephaly in neonatal rats depend on the pattern of alcohol administration. Alcohol 5, 209–214https://doi.org/10.1016/0741-8329(88)90054-7 [DOI] [PubMed] [Google Scholar]
- 9.Kietzman H. W., Everson J. L., Sulik K. K., Lipinski R. J. (2014) The teratogenic effects of prenatal ethanol exposure are exacerbated by Sonic Hedgehog or GLI2 haploinsufficiency in the mouse. PLoS One 9, e89448.https://doi.org/10.1371/journal.pone.0089448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCarthy N., Wetherill L., Lovely C. B., Swartz M. E., Foroud T. M., Eberhart J. K. (2013) Pdgfra protects against ethanol-induced craniofacial defects in a zebrafish model of FASD. Development 140, 3254–3265https://doi.org/10.1242/dev.094938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramanathan R., Wilkemeyer M. F., Mittal B., Perides G., Charness M. E. (1996) Alcohol inhibits cell-cell adhesion mediated by human L1. J. Cell Biol. 133, 381–390https://doi.org/10.1083/jcb.133.2.381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bearer C. F., Swick A. R., O’Riordan M. A., Cheng G. (1999) Ethanol inhibits L1-mediated neurite outgrowth in postnatal rat cerebellar granule cells. J. Biol. Chem. 274, 13264–13270https://doi.org/10.1074/jbc.274.19.13264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arevalo E., Shanmugasundararaj S., Wilkemeyer M. F., Dou X., Chen S., Charness M. E., Miller K. W. (2008) An alcohol binding site on the neural cell adhesion molecule L1. Proc. Natl. Acad. Sci. USA 105, 371–375https://doi.org/10.1073/pnas.0707815105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilkemeyer M. F., Charness M. E. (1998) Characterization of ethanol-sensitive and insensitive fibroblast cell lines expressing human L1. J. Neurochem. 71, 2382–2391https://doi.org/10.1046/j.1471-4159.1998.71062382.x [DOI] [PubMed] [Google Scholar]
- 15.Buhusi M., Midkiff B. R., Gates A. M., Richter M., Schachner M., Maness P. F. (2003) Close homolog of L1 is an enhancer of integrin-mediated cell migration. J. Biol. Chem. 278, 25024–25031https://doi.org/10.1074/jbc.M303084200 [DOI] [PubMed] [Google Scholar]
- 16.Dou X., Menkari C. E., Shanmugasundararaj S., Miller K. W., Charness M. E. (2011) Two alcohol binding residues interact across a domain interface of the L1 neural cell adhesion molecule and regulate cell adhesion. J. Biol. Chem. 286, 16131–16139https://doi.org/10.1074/jbc.M110.209254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen M. M., Lee C. Y., Leland H. A., Lin G. Y., Montgomery A. M., Silletti S. (2010) Inside-out regulation of L1 conformation, integrin binding, proteolysis, and concomitant cell migration. Mol. Biol. Cell 21, 1671–1685https://doi.org/10.1091/mbc.E09-10-0900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schaefer A. W., Kamei Y., Kamiguchi H., Wong E. V., Rapoport I., Kirchhausen T., Beach C. M., Landreth G., Lemmon S. K., Lemmon V. (2002) L1 endocytosis is controlled by a phosphorylation-dephosphorylation cycle stimulated by outside-in signaling by L1. J. Cell Biol. 157, 1223–1232https://doi.org/10.1083/jcb.200203024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schneider C. A., Rasband W. S., Eliceiri K. W. (2012) NIH image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675https://doi.org/10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilkemeyer M. F., Pajerski M., Charness M. E. (1999) Alcohol inhibition of cell adhesion in BMP-treated NG108-15 cells. Alcohol. Clin. Exp. Res. 23, 1711–1720https://doi.org/10.1111/j.1530-0277.1999.tb04065.x [PubMed] [Google Scholar]
- 21.Suttie M., Foroud T., Wetherill L., Jacobson J. L., Molteno C. D., Meintjes E. M., Hoyme H. E., Khaole N., Robinson L. K., Riley E. P., Jacobson S. W., Hammond P. (2013) Facial dysmorphism across the fetal alcohol spectrum. Pediatrics 131, e779–e788https://doi.org/10.1542/peds.2012-1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ignelzi M. A., Jr., Miller D. R., Soriano P., Maness P. F. (1994) Impaired neurite outgrowth of src-minus cerebellar neurons on the cell adhesion molecule L1. Neuron 12, 873–884https://doi.org/10.1016/0896-6273(94)90339-5 [DOI] [PubMed] [Google Scholar]
- 23.Chen S., Charness M. E. (2008) Ethanol inhibits neuronal differentiation by disrupting activity-dependent neuroprotective protein signaling. Proc. Natl. Acad. Sci. USA 105, 19962–19967https://doi.org/10.1073/pnas.0807758105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wong E. V., Schaefer A. W., Landreth G., Lemmon V. (1996) Involvement of p90rsk in neurite outgrowth mediated by the cell adhesion molecule L1. J. Biol. Chem. 271, 18217–18223https://doi.org/10.1074/jbc.271.30.18217 [DOI] [PubMed] [Google Scholar]
- 25.Wong E. V., Schaefer A. W., Landreth G., Lemmon V. (1996) Casein kinase II phosphorylates the neural cell adhesion molecule L1. J. Neurochem. 66, 779–786https://doi.org/10.1046/j.1471-4159.1996.66020779.x [DOI] [PubMed] [Google Scholar]
- 26.Davis J. Q., Bennett V. (1994) Ankyrin binding activity shared by the neurofascin/L1/NrCAM family of nervous system cell adhesion molecules. J. Biol. Chem. 269, 27163–27166 [PubMed] [Google Scholar]
- 27.Garver T. D., Ren Q., Tuvia S., Bennett V. (1997) Tyrosine phosphorylation at a site highly conserved in the L1 family of cell adhesion molecules abolishes ankyrin binding and increases lateral mobility of neurofascin. J. Cell Biol. 137, 703–714https://doi.org/10.1083/jcb.137.3.703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tuvia S., Garver T. D., Bennett V. (1997) The phosphorylation state of the FIGQY tyrosine of neurofascin determines ankyrin-binding activity and patterns of cell segregation. Proc. Natl. Acad. Sci. USA 94, 12957–12962https://doi.org/10.1073/pnas.94.24.12957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dai J., Buhusi M., Demyanenko G. P., Brennaman L. H., Hruska M., Dalva M. B., Maness P. F. (2013) Neuron glia-related cell adhesion molecule (NrCAM) promotes topographic retinocollicular mapping. PLoS One 8, e73000.https://doi.org/10.1371/journal.pone.0073000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang X., Davis J. Q., Carpenter S., Bennett V. (1998) Structural requirements for association of neurofascin with ankyrin. J. Biol. Chem. 273, 30785–30794https://doi.org/10.1074/jbc.273.46.30785 [DOI] [PubMed] [Google Scholar]
- 31.Needham L. K., Thelen K., Maness P. F. (2001) Cytoplasmic domain mutations of the L1 cell adhesion molecule reduce L1-ankyrin interactions. J. Neurosci. 21, 1490–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fransen E., Van Camp G., D’Hooge R., Vits L., Willems P. J. (1998) Genotype-phenotype correlation in L1 associated diseases. J. Med. Genet. 35, 399–404https://doi.org/10.1136/jmg.35.5.399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fransen E., Lemmon V., Van Camp G., Vits L., Coucke P., Willems P. J. (1995) CRASH syndrome: clinical spectrum of corpus callosum hypoplasia, retardation, adducted thumbs, spastic paraparesis and hydrocephalus due to mutations in one single gene, L1. Eur. J. Hum. Genet. 3, 273–284https://doi.org/10.1159/000472311 [DOI] [PubMed] [Google Scholar]
- 34.De Angelis E., Watkins A., Schäfer M., Brümmendorf T., Kenwrick S. (2002) Disease-associated mutations in L1 CAM interfere with ligand interactions and cell-surface expression. Hum. Mol. Genet. 11, 1–12https://doi.org/10.1093/hmg/11.1.1 [DOI] [PubMed] [Google Scholar]
- 35.Zhao X., Siu C. H. (1996) Differential effects of two hydrocephalus/MASA syndrome-related mutations on the homophilic binding and neuritogenic activities of the cell adhesion molecule L1. J. Biol. Chem. 271, 6563–6566https://doi.org/10.1074/jbc.271.12.6563 [DOI] [PubMed] [Google Scholar]
- 36.Chen S.-Y., Wilkemeyer M. F., Sulik K. K., Charness M. E. (2001) Octanol antagonism of ethanol teratogenesis. FASEB J. 15, 1649–1651 [DOI] [PubMed] [Google Scholar]
- 37.Wilkemeyer M. F., Chen S. Y., Menkari C. E., Brenneman D. E., Sulik K. K., Charness M. E. (2003) Differential effects of ethanol antagonism and neuroprotection in peptide fragment NAPVSIPQ prevention of ethanol-induced developmental toxicity. Proc. Natl. Acad. Sci. USA 100, 8543–8548https://doi.org/10.1073/pnas.1331636100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen S. Y., Charness M. E., Wilkemeyer M. F., Sulik K. K. (2005) Peptide-mediated protection from ethanol-induced neural tube defects. Dev. Neurosci. 27, 13–19https://doi.org/10.1159/000084528 [DOI] [PubMed] [Google Scholar]
- 39.Vallejo Y., Hortsch M., Dubreuil R. R. (1997) Ethanol does not inhibit the adhesive activity of Drosophila neuroglian or human L1 in Drosophila S2 tissue culture cells. J. Biol. Chem. 272, 12244–12247https://doi.org/10.1074/jbc.272.18.12244 [DOI] [PubMed] [Google Scholar]
- 40.Dai J., Dalal J. S., Thakar S., Henkemeyer M., Lemmon V. P., Harunaga J. S., Schlatter M. C., Buhusi M., Maness P. F. (2012) EphB regulates L1 phosphorylation during retinocollicular mapping. Mol. Cell. Neurosci. 50, 201–210https://doi.org/10.1016/j.mcn.2012.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dou X., Charness M. E. (2014) Effect of lipid raft disruption on ethanol inhibition of l1 adhesion. Alcohol. Clin. Exp. Res. 38, 2707–2711https://doi.org/10.1111/acer.12556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bennett V., Lambert S. (1999) Physiological roles of axonal ankyrins in survival of premyelinated axons and localization of voltage-gated sodium channels. J. Neurocytol. 28, 303–318https://doi.org/10.1023/A:1007005528505 [DOI] [PubMed] [Google Scholar]
- 43.Hortsch M., Homer D., Malhotra J. D., Chang S., Frankel J., Jefford G., Dubreuil R. R. (1998) Structural requirements for outside-in and inside-out signaling by Drosophila neuroglian, a member of the L1 family of cell adhesion molecules. J. Cell Biol. 142, 251–261https://doi.org/10.1083/jcb.142.1.251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haspel J., Grumet M. (2003) The L1CAM extracellular region: a multi-domain protein with modular and cooperative binding modes. Front. Biosci. 8, s1210–s1225https://doi.org/10.2741/1108 [DOI] [PubMed] [Google Scholar]
- 45.Yamasaki M., Thompson P., Lemmon V. (1997) CRASH syndrome: mutations in L1CAM correlate with severity of the disease. Neuropediatrics 28, 175–178https://doi.org/10.1055/s-2007-973696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kizhatil K., Davis J. Q., Davis L., Hoffman J., Hogan B. L., Bennett V. (2007) Ankyrin-G is a molecular partner of E-cadherin in epithelial cells and early embryos. J. Biol. Chem. 282, 26552–26561https://doi.org/10.1074/jbc.M703158200 [DOI] [PubMed] [Google Scholar]
- 47.Hoffman E. J., Mintz C. D., Wang S., McNickle D. G., Salton S. R., Benson D. L. (2008) Effects of ethanol on axon outgrowth and branching in developing rat cortical neurons. Neuroscience 157, 556–565https://doi.org/10.1016/j.neuroscience.2008.08.071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Iqbal Z., Vandeweyer G., van der Voet M., Waryah A. M., Zahoor M. Y., Besseling J. A., Roca L. T., Vulto-van Silfhout A. T., Nijhof B., Kramer J. M., Van der Aa N., Ansar M., Peeters H., Helsmoortel C., Gilissen C., Vissers L. E., Veltman J. A., de Brouwer A. P., Frank Kooy R., Riazuddin S., Schenck A., van Bokhoven H., Rooms L. (2013) Homozygous and heterozygous disruptions of ANK3: at the crossroads of neurodevelopmental and psychiatric disorders. Hum. Mol. Genet. 22, 1960–1970https://doi.org/10.1093/hmg/ddt043 [DOI] [PubMed] [Google Scholar]
- 49.Jenkins P. M., Kim N., Jones S. L., Tseng W. C., Svitkina T. M., Yin H. H., Bennett V. (2015) Giant ankyrin-G: a critical innovation in vertebrate evolution of fast and integrated neuronal signaling. Proc. Natl. Acad. Sci. USA 112, 957–964https://doi.org/10.1073/pnas.1416544112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ota M., Hori H., Sato N., Yoshida F., Hattori K., Teraishi T., Kunugi H. (2016) Effects of ankyrin 3 gene risk variants on brain structures in patients with bipolar disorder and healthy subjects. Psychiatry Clin. Neurosci. 70, 498–506https://doi.org/10.1111/pcn.12431 [DOI] [PubMed] [Google Scholar]
- 51.Bi C., Wu J., Jiang T., Liu Q., Cai W., Yu P., Cai T., Zhao M., Jiang Y. H., Sun Z. S. (2012) Mutations of ANK3 identified by exome sequencing are associated with autism susceptibility. Hum. Mutat. 33, 1635–1638https://doi.org/10.1002/humu.22174 [DOI] [PubMed] [Google Scholar]
- 52.Zeniou M., Ding T., Trivier E., Hanauer A. (2002) Expression analysis of RSK gene family members: the RSK2 gene, mutated in Coffin-Lowry syndrome, is prominently expressed in brain structures essential for cognitive function and learning. Hum. Mol. Genet. 11, 2929–2940https://doi.org/10.1093/hmg/11.23.2929 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.