Figure 5. CONCR Interacts with DDX11 Protein and Regulates Its Function.
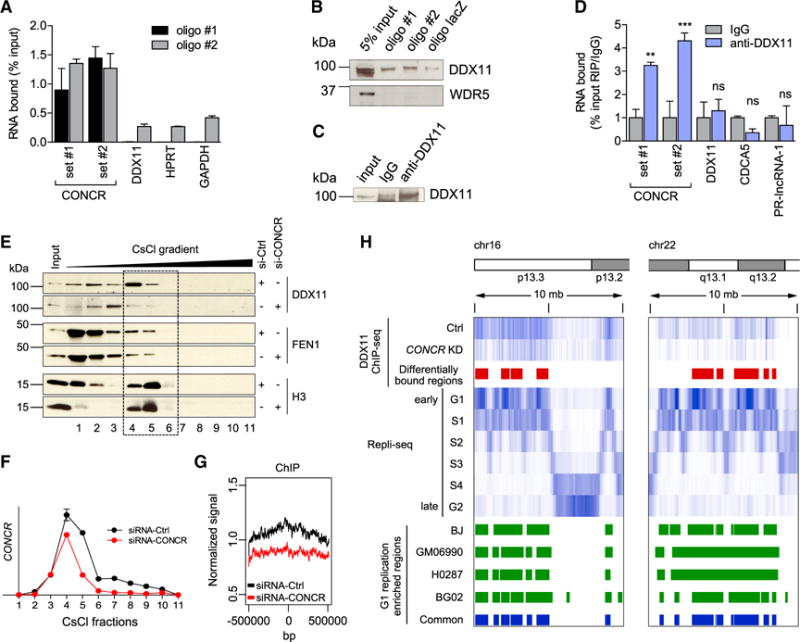
(A) Percentage of CONCR and control RNAs (DDX11, HPRT, and GAPDH) pulled down with two independent biotinylated oligonucleotides (oligo #1 and oligo #2) in A549 cells.
(B) DDX11 and WDR5 (control) western blots of the CONCR associated proteins pulled down as in (A).
(C) Western blot of DDX11 immunoprecipitation as in (D).
(D) Percentage of CONCR and control RNAs (DDX11, CDCA5, and PR-lncRNA-1) immunoprecipitated with an anti-DDX11 antibody or IgG control. Graph shows mean ± SD of three independent experiments. Significance was determined comparing to IgG.
(E) DDX11, FEN1, and histone 3 (H3) distribution in the fractions obtained from CsCl density-gradient centrifugation of A549 cells transfected with control or CONCR siRNAs as indicated.
(F) Quantification by qRT-PCR of CONCR in the gradient fractions shown in (E).
(G) Mean DDX11 ChIP-seq signal around the center of all the genomic regions identified as differentially bound by DDX11 in CONCR-depleted cells compared to siRNA-Ctrl-transfected cells.
(H) DDX11 ChIP-seq. Top to bottom: chromosome schematic of two representative regions of chr16 and chr22; DDX11 ChIP-seq signal in control (Ctrl) and CONCR-depleted cells (KD); regions with differential binding of DDX11 (Ctrl versus KD); signals of DNA replicating regions of BJ cells in G1 to G2 phases of the cell cycle as reported in Hansen et al. (2010); G1-replication-enriched regions common to BJ, GM06990, H0287, and BG02 cell types as reported in Hansen et al. (2010).
See also Figures S5 and S6, and Table S5.
