Figure 3. InR and its downstream pathways in ACs are important for the starvation-induced reduction in Tp.
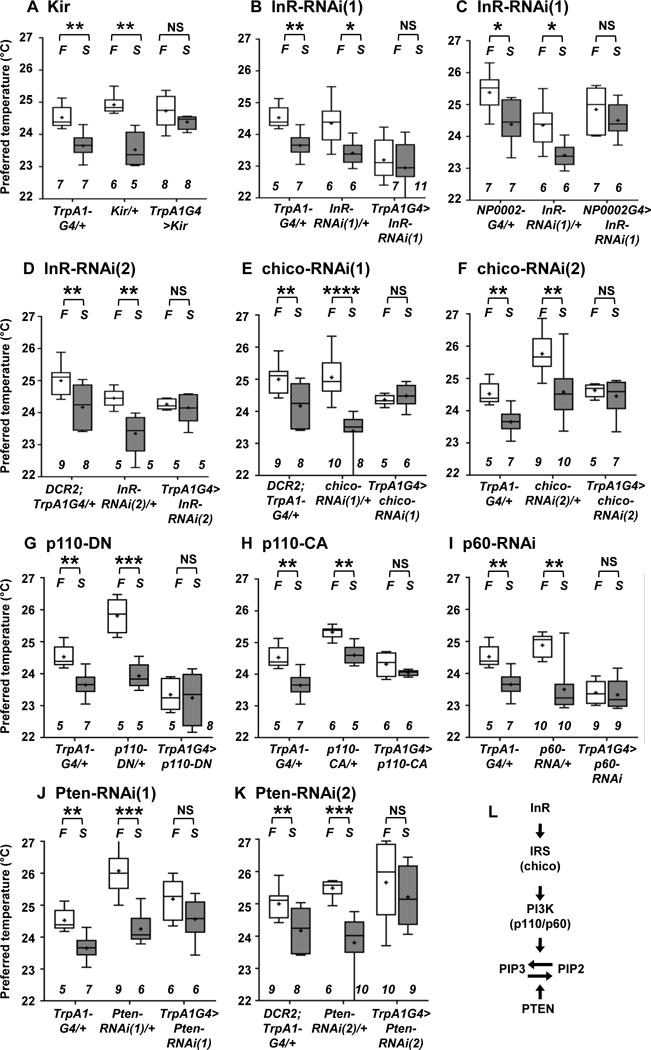
Comparison of Tp between the fed (F: fed, white box) and starved overnight (S: starved O/N, gray box) conditions in the groups of flies indicated below.
(A) Inhibition of ACs by Kir expression: TrpA1G4/+, Kir/+ (UAS-Kir/+) and TrpA1G4>Kir (TrpA1SH-Gal4/+; UAS-Kir/+) flies. AC-inhibited flies still avoided warm temperatures and preferred a temperature of ~25 °C in fed conditions (white box). These data suggest that ACs may not be the only neurons that regulate warmth-avoidance behavior and that other redundant temperature-sensing mechanisms could contribute to this warm avoidance phenotype under the conditions applied in the present study. Given that ACs are responsible for the starvation-induced reduction in Tp, ACs represent the critical thermosensor for setting a new homeostatic set-point in starved conditions but might not be sole regulator of the warmth-avoidance phenotype.
(B) RNAi knockdown of InR in ACs: TrpA1G4/+, InR-RNAi(1)/+ (UAS- InR-RNAi(1)/+) and TrpA1G4>InR-RNAi(1) (TrpA1SH-Gal4/UAS-InR-RNAi(1)) flies.
(C) RNAi knockdown of InR in ACs: NP0002/+ (NP0002-Gal4/+), InR-RNAi(1)/+ (UAS- InR-RNAi(1)/+) and NP0002>InR-RNAi(1) (NP0002-Gal4/UAS-InR-RNAi(1)) flies.
(D) RNAi knockdown of InR in ACs: DCR2; TrpA1G4/+ (uas-DCR2/+; TrpA1SH-Gal4/+), InR-RNAi(2)/+ (uas-InR-RNAi(2)/+) and DCR2; TrpA1G4> InR-RNAi(2) (uas-DCR2/+; TrpA1SH-Gal4/+; uas-InR-RNAi(2)/+) flies. InR-RNAi(1) and InR-RNAi(2) are different RNAi lines targeting different regions of the InR sequence.
(E) RNAi knockdown of chico in ACs: DCR2; TrpA1G4/+, chico-RNAi(1)/+ (uas-chico-RNAi(1)/+) and TrpA1G4> chico-RNAi(1) (uas-DCR2/+; TrpA1SH-Gal4/+; uas-chico-RNAi(1)/+) flies.
(F) RNAi knockdown of chico in ACs: TrpA1G4/+, chico-RNAi(2)/+ (uas-chico-RNAi(2)/+) and TrpA1G4> chico-RNAi(2) (TrpA1SH-Gal4/uas-chico-RNAi(2)) flies. chico-RNAi(1) and chico-RNAi(2) are different RNAi lines targeting different regions of the chico sequence.
(G) Expression of a dominant-negative form of p110 (p110-DN) in ACs: TrpA1G4/+, p110-DN/+ (uas-p110-DN/+) and TrpA1G4>p110-DN (TrpA1SH-Gal4/uas-p110-DN) flies. DN means a dominant-negative form of p110.
(H) Expression of a constitutive active form of p110 (p110-CA) in ACs: TrpA1G4/+, p110-CA/+ (uas-p110-CA/+) and TrpA1G4>p110-CA (uas-p110-CA/+; TrpA1SH-Gal4/+) flies. CA means a constitutive active form of p110.
(I) RNAi knockdown of p60 in ACs: TrpA1G4/+, p60-RNAi/+ (uas-p60-RNAi/+) and TrpA1G4>p60-RNAi (TrpA1SH-Gal4/uas-p60-RNAi) flies.
(J) RNAi knockdown of Pten in ACs: TrpA1G4/+, Pten-RNAi (1)/+ (uas-Pten-RNAi(1)/+) and TrpA1G4>Pten-RNAi (1) (TrpA1SH-Gal4/uas-Pten-RNAi(1)) flies.
(K) RNAi knockdown of Pten in ACs: TrpA1G4/+, Pten-RNAi(2)/+ (uas-Pten-RNAi(2)/+) and TrpA1G4> Pten-RNAi(1) (uas-DCR2/+; TrpA1SH-Gal4/+; uas-Pten-RNAi(2)/+) flies. Pten-RNAi(1) and Pten-RNAi(2) are different RNAi lines targeting different regions of the Pten sequence.
(L) A schematic diagram of IIS.
The plotting pattern and statistical analysis are the same as in Figure 2A. *p<0.05. **p<0.01. ***p<0.001. ****p<0.0001. NS indicates no significance. See also Figure S4, Table S1 and S2.
