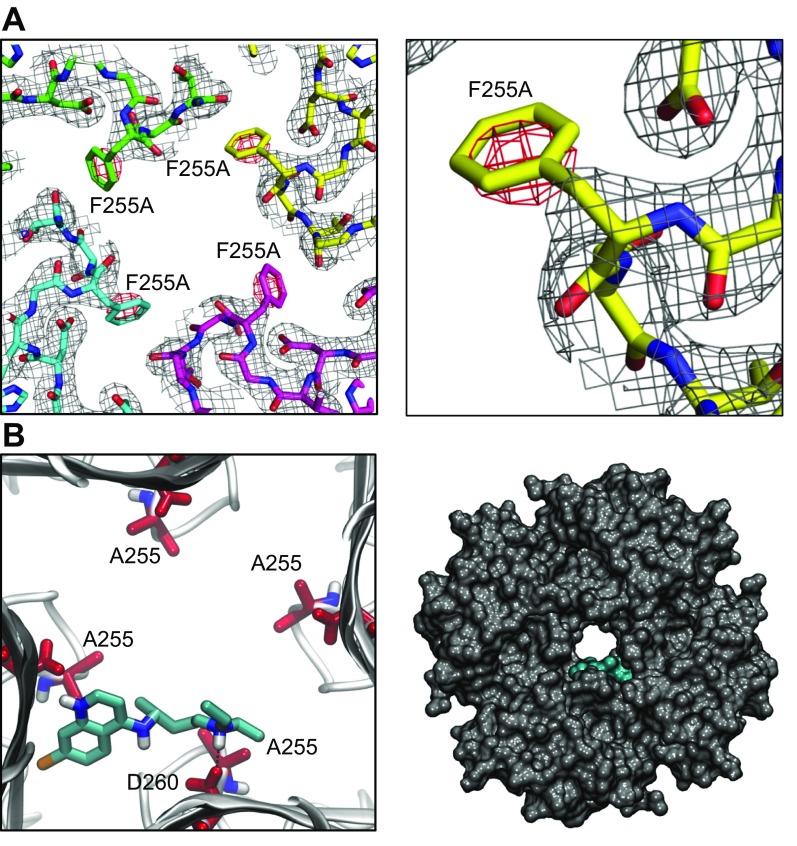
Figure 6.
X-Ray crystallography structure of Kir3.1 with F255A mutation at 2.5 Å resolution and docking of chloroquine in the mutant channel structure. A) Left: electron density map for Kir3.1 F255A tetrameric channel after molecular replacement. The difference (Fo-Fc) electron density map, around the area of residue 255, is contoured in red at −3 σ. Right: magnified view of the residue 255 region from 1 subunit highlighting the red negative-density blob, which indicates the absence of residue F255. B) Docking of chloroquine into the ion-permeation pathway of the F255A mutant Kir3.1 channel. Left: magnified view of the binding pose of chloroquine (cyan sticks) in Kir3.1. Chloroquine binding is off centered, with the amine nitrogen of chloroquine forming a hydrogen bond with residue D260. The F255 residues from each of the 4 Kir3.1 subunits are shown in red sticks. Right) van der Waals representation of the F255A mutant channel bound to chloroquine (cyan), viewed from the intracellular side.