FIGURE 4.
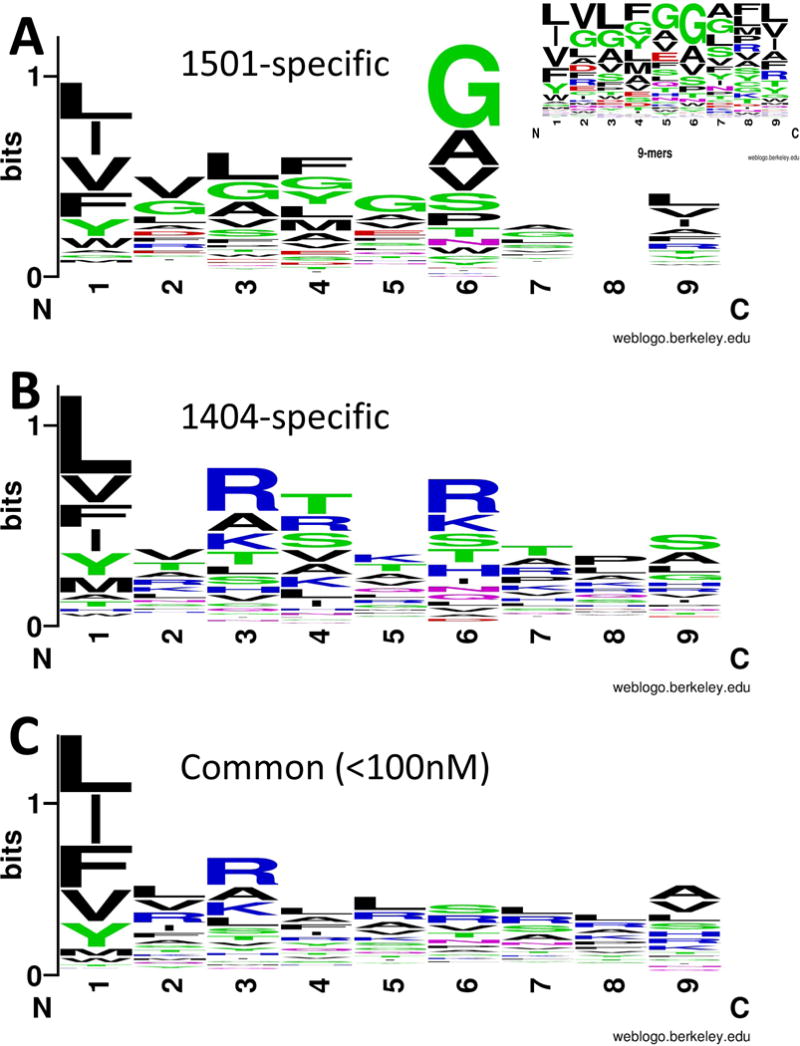
Sequence motifs that characterise 9-mer core epitopes for 20-mer peptides predicted to bind preferentially to (A) DRB1*1501 or (B) DRB1*1404 alleles, or (C) predicted to bind to both DRB1 alleles at <100nM. Preferential binders were defined as those with ≤500nM affinity for one DRB1 allele and ≥1000nM for the alternative allele (see Table S1 for examples). In silico predictions for 20-mer peptides were generated across 49 Leishmania vaccine candidate proteins using NetMHCIIpan 2.1. Sequences were aligned using BlockLogo and plotted using WebLogo, as referenced in the main text. The plots are based on 9-mer cores for 20-mer epitopes that spanned the peaks as plotted in Figure S1. Only one 9-mer core was selected to represent each epitope peak. The colours of the amino acids (AA) correspond to their chemical properties: polar AA are shown in green (G,S,T,Y,C; note G is non-polar when it forms a bond with other AA) or purple (Q,N); basic AA in blue (K,R,H); acidic AA in red (D,E); hydrophobic AA in black (A,V,L,I,P,W,F,M). The size of the letter relates to the frequency with which the AA is found in this position of core 9-mers, the overall peak height on the y-axis indicates the degree of conservation for specific epitopes at this location. Position 8 for DRB1*1501 showed no AA conservation; frequencies of AA at this position are shown in the insert figure.
