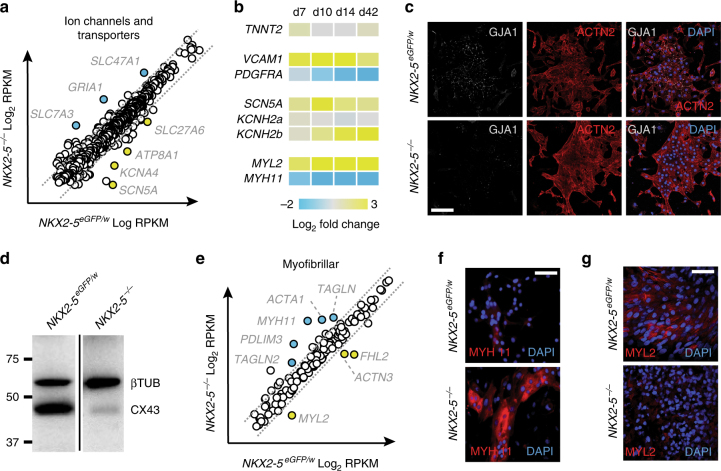
Fig. 4.
Deletion of NKX2-5 disrupts both electrical and mechanical gene networks in cardiomyocytes. a Dot plot representation of RNA-seq absolute gene expression (log2 RPKM values) for a reported list of ion channel and transporter genes. Dotted lines mark 2-fold differential expression level. b Heat map of gene expression (Q-PCR) in GFP+ cells isolated from NKX2-5eGFP/w and NKX2-5−/− cultures at day 7, 10, 14, or 42 of cardiac differentiation. Displayed as mean log2 fold change between the two genotypes at each time point (n = 4). c Immunocytochemistry analysis of GJA1 (CX43) and ACTN2 expression in cardiomyocytes derived from NKX2-5eGFP/w and NKX2-5−/− cells at day 42 of differentiation. Scale bar = 100 μm. d Western blot detection of GJA1 in NKX2-5eGFP/w and NKX2-5−/− cultures confirms reduction in GJA1 observed in h. Size markers in kDa are indicated to the left of the blot. e Dot plot representation of RNA-seq absolute gene expression (log2 RPKM values) for myofibrillar genes. Dotted lines mark 2-fold differential expression level. f Immunofluorescent detection of MYH11 (smooth muscle myosin heavy chain) in cardiomyocytes derived from NKX2-5eGFP/w and NKX2-5−/− cells at day 14 of differentiation. Nuclei are counterstained with DAPI. Scale bar = 50 μm. g Immunofluorescent detection of MYL2 (Myosin light chain 2 v) in cardiac organoids generated from NKX2-5eGFP/w and NKX2-5−/− cells. Nuclei are counterstained with DAPI. Scale bar = 50 μm