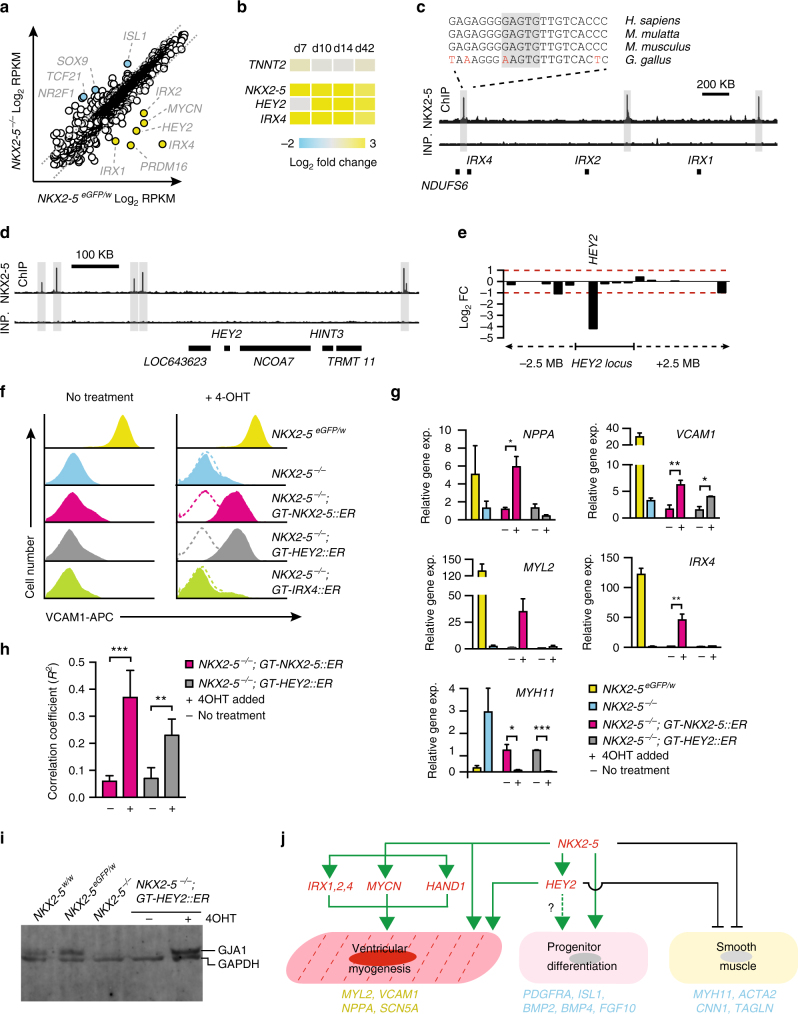
Fig. 5.
HEY2 is a key downstream transcriptional mediator of NKX2-5. a Dot plot representation of RNA-seq absolute gene expression (log2 RPKM values) for FANTOM5 predicted transcription factors. Dotted line marks 2 fold differential expression level. b Heat map of gene expression in GFP+ cells isolated from NKX2-5eGFP/w and NKX2-5−/− cultures at day 7, 10 or 14 of cardiac differentiation, as determined by Q-PCR. Displayed as mean log2 fold change between the two genotypes at each time point (n = 4). c, d Schematics of NKX2-5 ChIP-seq data showing the IRX1/2/4 cluster (c) and HEY2 locus (d) with regions bound by NKX2-5 highlighted. The IRX4 proximal NKX2-5 bound region is highly conserved. Inp. = input chromatin. e Differential expression of genes 2.5 Mbp up or downstream of the HEY2 locus in d. This data shows HEY2 is the only differentially expressed gene in this chromosomal region. Red dashed line marks 2 fold (adj. p value < 0.05) gene expression difference between genotypes. f Histograms of flow cytometry analysis of VCAM1 in untreated (No treatment) or induced (+4-OHT) NKX2-5−/− GAPTrap (GT) lines. Both GT-NKX2-5::ER and GT-HEY::ER restore VCAM1 expression (n = 4). g Gene expression profiling of genetic rescue via the modified GAPTrap loci, GT-NKX2-5::ER, GT-HEY::ER and GT-IRX4::ER, as determined by Q-PCR (n = 3). * p < 0.05, ** p < 0.01, *** p < 0.001 (Student’s t-test). h Correlation coefficient between contractile areas improves when both NKX2-5 and HEY2 are induced (n = 3, scored blind to genotype). ** p < 0.01, *** p < 0.001 (Student’s t-test). i Western blot showing restoration of GJA1 (connexin 43) levels by HEY2 and that wildtype (HES3) and NKX2-5eGFP/w GJA1 levels are comparable. j Network model of NKX2-5 regulated genes and their potential roles in regulating ventricular myogenesis, progenitor differentiation and smooth muscle differentiation. Representative genes with altered expression (yellow text activated genes, blue repressed genes) in NKX2-5 null cultures are shown below each process