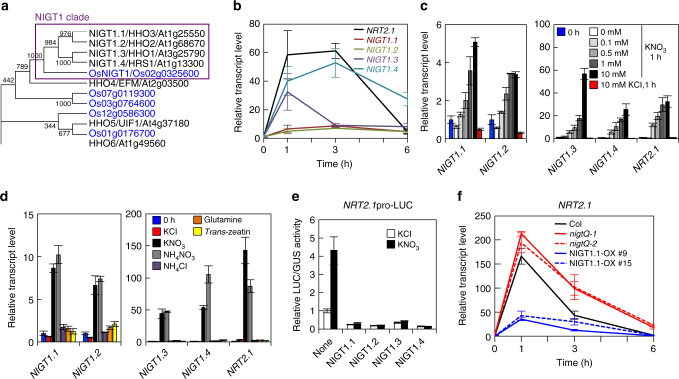
Fig. 2.
NIGT1 proteins redundantly regulate NRT2.1 expression. a Phylogenic analysis of Arabidopsis and rice NIGT1/HHO proteins. Arabidopsis and rice proteins are shown in black and blue, respectively. Numbers indicate bootstrap values of 1000 iterations. b Time-dependent expression patterns of NIGT1-clade genes after 10 mM KNO3 supply. c Concentration-dependent induction by nitrate. RNA was prepared from ammonium-grown seedlings before treatment and after treatment with 0, 0.1, 0.5, 1 and 10 mM KNO3 for 1 h. A 10 mM KCl treatment was used as a control. d Nitrate-specific induction of NIGT1-clade genes. Ammonium-grown seedlings were treated for 1 h with 10 mM of each chemical, except for trans-zeatin (5 µM). e Repression of the NRT2.1 promoter by NIGT1 proteins. Protoplasts co-transfected with NIGT1 expression vectors or the empty vector (none) together with the reporter plasmid containing the LUC gene under the control of the NRT2.1 promoter were incubated in the presence of 1 mM KCl or KNO3. LUC activity was normalised to GUS activity from the co-transfected UBQ10-GUS plasmid. Data are means ± s.d. of three biological replicates. f Transcript levels of NRT2.1 in the nigt1 quadruple mutants (Q-1 and Q-2) and NIGT1.1-OX plants after 10 mM KNO3 supply. Values are normalised to those of UBQ10, and are means of biological triplicates ± s.d. (b, c, d, f)