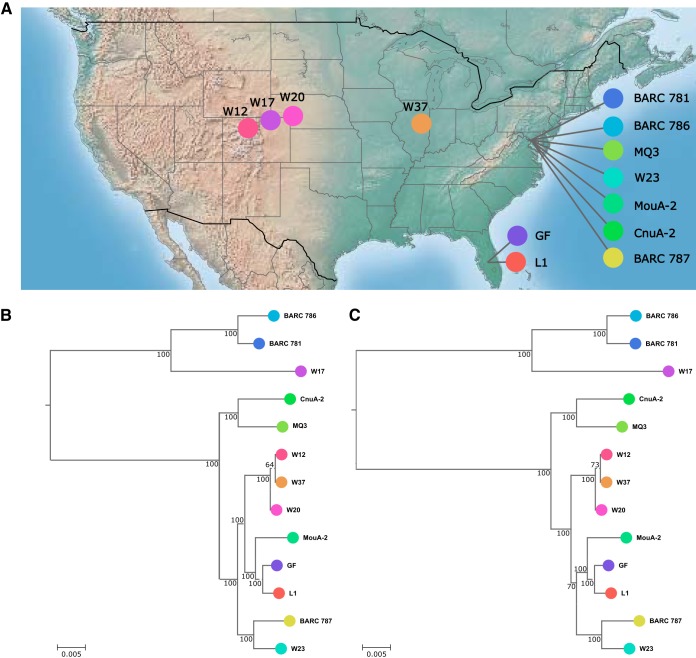
FIG 1 .
M. florum strain sampling and phylogeny. (A) Isolation sites of the 13 M. florum strains analyzed in this study. (B, C) M. florum phylogenetic trees constructed by using concatenated alignments of 412 conserved proteins and the Kimura distance model (B) or maximum likelihood (C). M. capricolum was used as the outgroup for both trees and is not shown because of the long branch length. Bootstrap values correspond to 100 repetitions. In both trees, branch length represents the substitution rate per site per unit of alignment length.