FIG 2 .
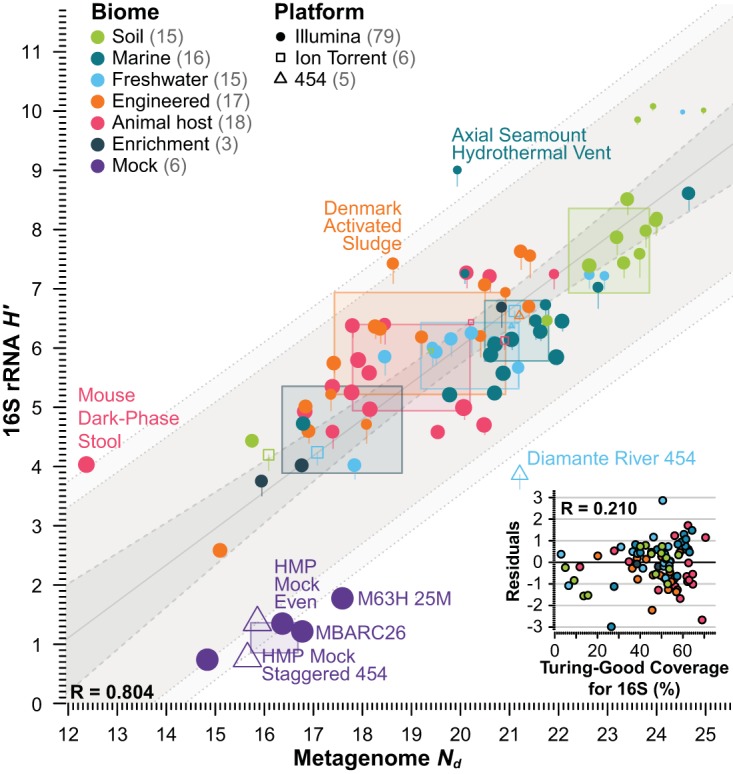
Comparison of Nonpareil Nd sequence diversity and 16S rRNA gene OTU Shannon H′ taxonomic diversity indices on 90 metagenomes. Each data point represents the estimates on Nd (x axis) and H′ (y axis). The y-axis value of each point indicates the Bayesian analysis-corrected Shannon index, and the line extending from the low part of each data point represents the exact observed (maximum-likelihood) Shannon index. The color of each point indicates the type of biome of each data set, the shape indicates the sequencing platform, and the size indicates the estimated coverage of the 16S rRNA gene profile (Turing-Good estimate). For each biome, the IQR of both estimates is represented as semitransparent rectangles. The least-squares linear correlation model is represented in gray, including the central estimate (solid line), the 95% confidence interval (dashed-line band), and the 80 and 95% prediction intervals (dotted-line bands). Labeled data sets fell outside the 80% prediction interval. The inset shows the residuals from the linear model against the Turing-Good estimate of 16S rRNA gene coverage.
