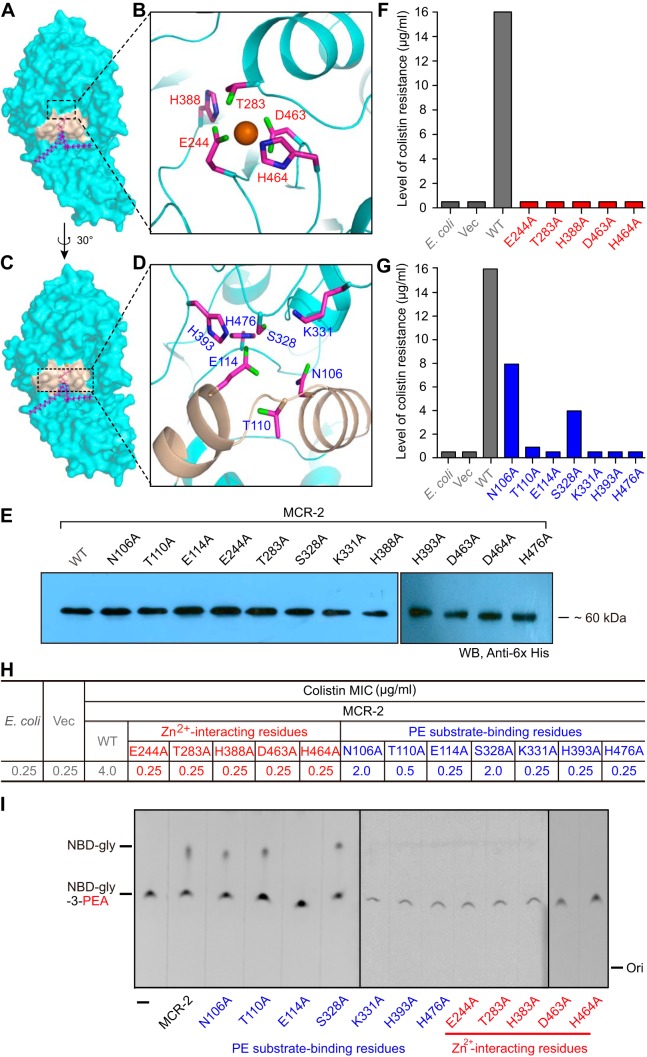
FIG 4 .
Structure-guided functional analyses of the PE-binding cavity of MCR-2. (A) Surface structure of MCR-2 with the cavity required for entry and binding of PE substrate. (B) Enlarged view of the 5-residue Zn2+-binding motif. (C) Surface structure of MCR-2 in the counter clockwise rotation of 30 degrees with fine-structural illustration of the cavity for entry of the PE substrate. (D) Enlarged view of the 7-residue motif with essential roles in the PE-binding cavity of MCR-2. The 7 amino acids (N106, T110, E114, S328, K331, H393, and H476) are proposed to participate in the formation of the PE cavity of MCR-2. (E) Western blotting-based expression analyses of MCR-2 and its 12 point-mutants in E. coli. Given the limit of the wells of PAGE (10 per gel), the photograph of Western blotting here was generated through a combination of two different gel images in which the protein samples were separated. (F) Site-directed mutagenesis assay of the Zn2+-binding motif in the context of MCR-2 colistin resistance. The 5 residues in the Zn2+-binding motif of MCR-2 are E244, T283, H388, D463, and H464. (G) Site-directed mutagenesis assay of the PE-binding cavity in the context of MCR-2 colistin resistance. The 2 periplasmic-facing helices (in light golden in panel D) possess 3 crucial residues (namely, N106, T110, and E114) that play roles in the binding of MCR-2 to the PE substrate molecule. (H) Comparison of colistin MICs in E. coli strains carrying either wild-type mcr-2 or its point mutants. (I) TLC-based assays of the enzymatic activities of MCR-2 and its 12 point mutants. Structure-guided site-directed mutagenesis was performed as recommended by the manufacturer. Vec, empty-vector-bearing strain; WT, wild type; Ori, origin. All strains tested here are listed in Table S1.