Fig. 3.
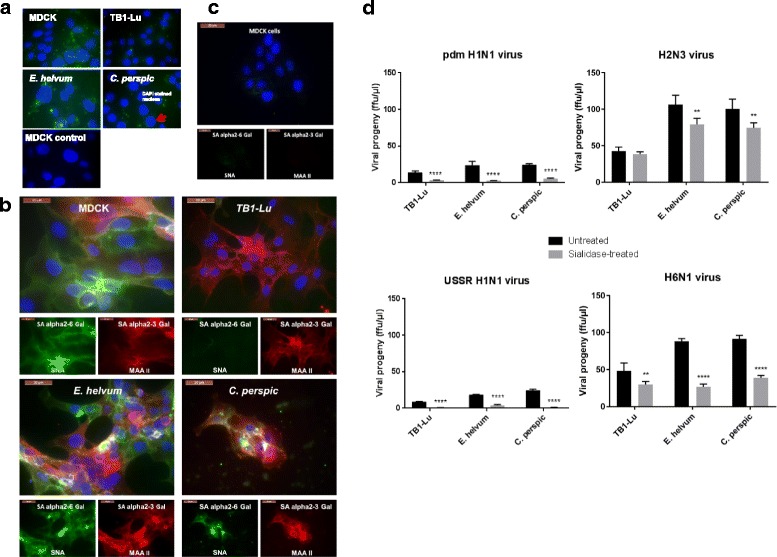
Host SA receptors contribute to influenza virus entry into bat cells. a TB1-Lu cells exhibited the least uptake of FITC-labelled siRNA in comparison with MDCK, E. helvum and C perspic cells. Respiratory epithelial cells from the three bat species and MDCK cells were transfected with FITC-labelled siRNA with the use of Viromer Blue reagent which mimics natural influenza viral entry and membrane fusion. Fluorescence imaging at 4 h showed detectable uptake of siRNA (green spots) in MDCK, E. helvum and C. perspic cells. However, TB1-Lu cells barely showed any fluorescence. Mock transfected MDCK cells showed complete absence of fluorescence. b E. helvum and C. persic but not TB1-Lu cells expressed human and avian SA receptors. Bat and MDCK cells were subjected to binding by plant lectins SNA and MAA II for the detection of human SAα2,6 Gal (green) and avian SAα2,3 Gal (red) receptors respectively. Avian SAα2,3 Gal and human SAα2,6 Gal receptors were readily found in all cell types except for TB1-Lu cells where SAα2,6 Gal receptor was barely detected. Nuclei were counterstained with DAPI (blue). c SA receptors were effectively removed from MDCK cells treated with a broad spectrum neuraminidase derived from Clostridium perfringens. Post-treatment, there was hardly any detection of SA receptors using MAA II and SNA lectins. d Collective removal of SA receptors with neuraminidase resulted in a marked reduction in progeny virus production from all three bat species. All results shown are representative of three experimental repeats
