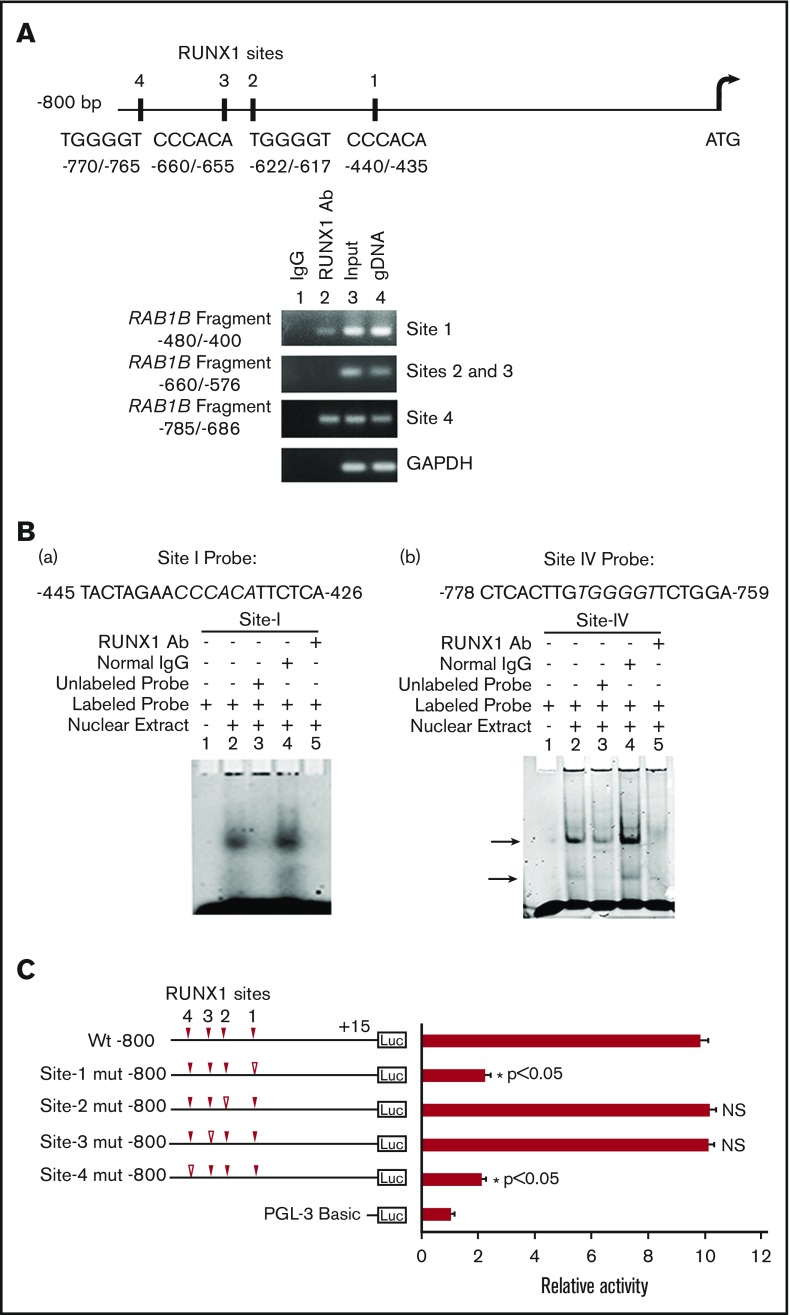
Figure 2.
Characterization of RUNX1 sites in the RAB1B upstream region. (A) RAB1B upstream region showing 4 consensus sites for RUNX1. Shown below is PCR amplification of the immunoprecipitates of HEL cells with control IgG (lane 1) and RUNX1 antibodies (lane 2). Amplification of the input or total DNA (lane 3) and genomic DNA (gDNA, lane 4) served as positive controls. Samples were analyzed by PCR using primers for the RAB1B region and GAPDH. Shown are representative of 3 experiments. (B) EMSA using WT nucleotide probes encompassing RUNX1 consensus site 1 (−426/−445; left) and site 4 (−759/−778; right) in RAB1B promoter and nuclear extracts from PMA-treated HEL cells. (a, left) EMSA using site 1 probe (lanes 1-5): lane 1, no extract; lane 2, protein binding to the probe; lane 3, loss of binding by competition with unlabeled probe; lane 4, no loss of binding by competition with normal IgG; and lane 5 competition with RUNX1 antibody and inhibition of binding. (b, right) EMSA using probe with site 4 (lanes 1-5); similar results were obtained as with the probe with site 1. Shown are representative of 3 experiments. (C) Luciferase reporter studies on RAB1B promoter in PMA-treated HEL cells. Luciferase activity with WT construct with RUNX1 sites 1-4 (solid triangles) and constructs with RUNX1 binding sites mutated (open triangles). Mutations in the sites 1 and 4 decreased promoter activity, but not the mutations in the sites 3 and 4, suggesting sites 1 and 4 are functional. The mean ± SEM is shown for 3 independent experiments in triplicates. P values are for comparisons against WT promoter.