Figure 5.
P. syringae Infection Triggers Proteaphagy.
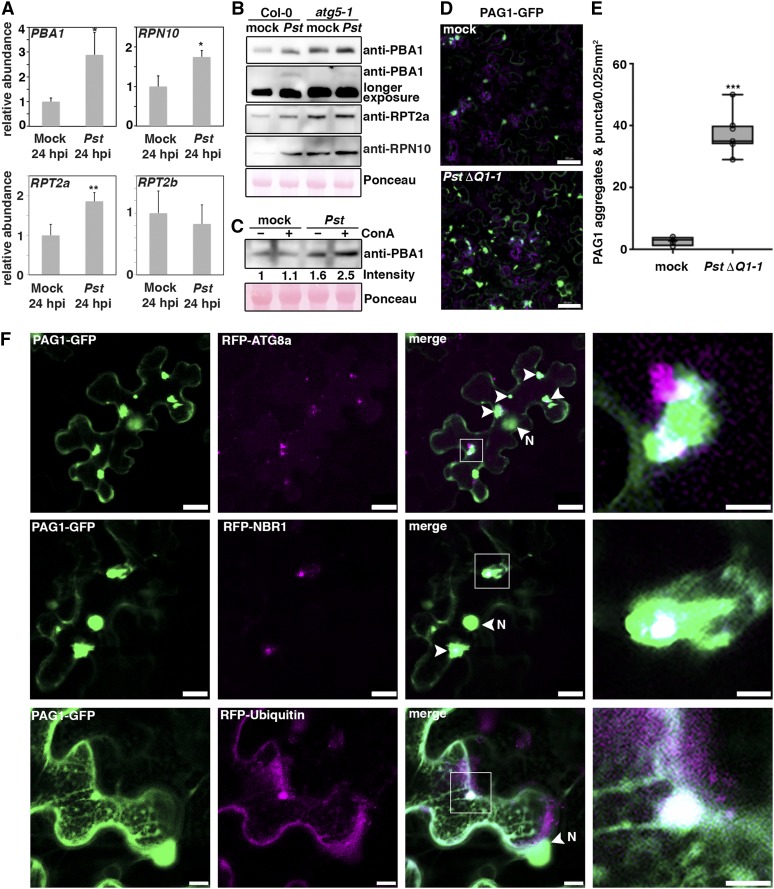
(A) RT-qPCR analysis of PBA1, RPN10, RPT2a, and RPT2b transcript levels upon challenge of Col-0 plants with Pst at 24 hpi compared with mock-infected plants. Values represent mean ± sd (n = 4) relative to the mock control and were normalized to PP2A. Statistical significance (*P < 0.05 and **P < 0.01) was revealed by Student’s t test (compared with mock control). The experiment was repeated twice with similar results.
(B) Immunoblot analysis of PBA1, RPT2a, and RPN10 protein levels in Col-0 and atg5-1 at 1 dpi with Pst. Proteins were extracted from leaves at 1 dpi and probed with specific antibodies. Ponceau S staining of RbcL served as a loading control. Immunoblots were repeated twice with similar results.
(C) Immunoblot analysis of PBA1 in the absence (–) and presence (+) of ConA during Pst infection. Proteins were extracted from leaves at 6 hpi and probed with specific antibodies. Numbers correspond to the relative signal intensities compared with the mock control without ConA. Ponceau S staining of RbcL was used as loading control and quantified for normalization of PBA1 signals. The experiment was repeated twice with similar results.
(D) Confocal analysis of transiently expressed PAG1-GFP in N. benthamiana at 6 hpi with PstΔhopQ1-1 compared with mock control. Images represent single confocal planes from abaxial epidermal cells and were acquired with identical microscope settings. The experiment was repeated at least three times with similar results. Bars = 50 μm.
(E) Quantification of PAG1-GFP aggregates in mock- and PstΔhopQ1-1-infected leaves analyzed in (C). The number of aggregates were calculated from single planes of independent leaf areas of 0.025 mm2 (n = 9) using ImageJ. Center lines show the medians; box limits indicate the 25th and 75th percentiles as determined by R software; whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles and data points are plotted as open circles. Asterisks indicate statistical significance (***P < 0.001) determined by Student’s t test in comparison with the control.
(F) Colocalization analysis of PAG1-GFP with RFP-ATG8a, RFP-NBR1, or RFP-Ub in N. benthamiana leaves. After 2 d of transient expression, plants were infected with PstΔhopQ1-1 to induce PAG1-GFP aggregates, and imaging was done at 4 to 6 hpi (bars = 20 μm). Arrows indicate areas of partial colocalization, and N marks the nucleus that also shows GFP fluorescence. Images in the right column are magnifications of boxed areas (bars = 5 μm). The experiment was repeated three times with similar results.