Figure 5.
Genetic Identification and Characterization of the LRR-RK ZmMSP1.
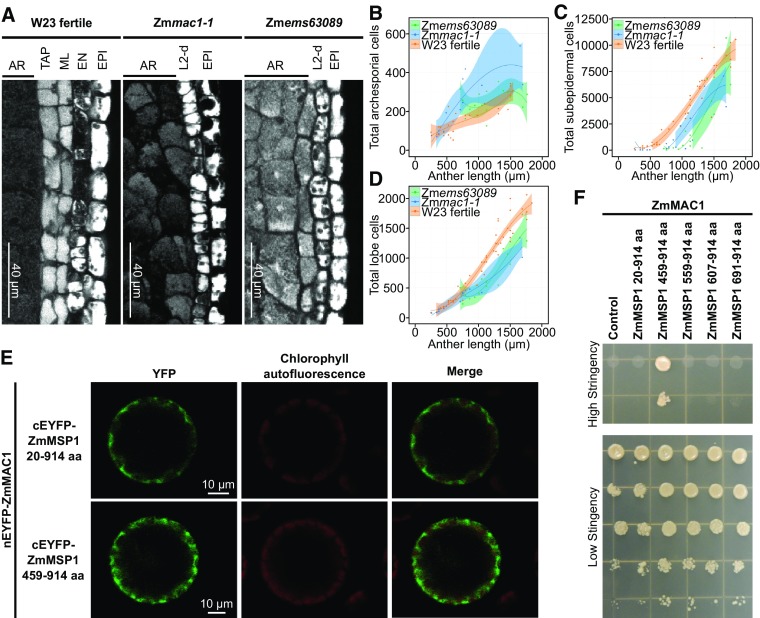
(A) Microscopy images of longitudinal sections of W23 fertile, Zmmac1-1, and Zmems63089 lobes stained with propidium iodide. EPI, epidermis cells.
(B) to (D) Plots of archesporial (B), L2-d or SPC +ML+ TAP (C), and total cells (D), per lobe in W23 fertile (orange) (Egger and Walbot, 2016), Zmems63089 (green) (Egger and Walbot, 2016), and Zmmac1-1 (blue). Data sets (dots) were fit to a third degree polynomial (line), and the shaded region is the 95% confidence interval (Egger and Walbot, 2016). Zmmac1-1 cell counts were performed on 21 anthers from two individual plants.
(E) BiFC assays of ZmMAC1 and ZmMSP1 fragments in Arabidopsis protoplasts. Part of the ZmMAC1 coding region was fused to nEYFP and cEFYP was fused to fragments of ZmMSP1. The resulting constructs were expressed in Arabidopsis protoplasts showing fluorescence complementation (left panel, EYFP shown in green) as a result of interaction. Intactness of protoplasts was visualized by chlorophyll autofluorescence (middle panel, red).
(F) Interaction study of ZmMAC1 and ZmMSP1 by yeast-two-hybrid assay. ZmMAC1 (bait) was tested in combination with the extracellular domain of ZmMSP1 (amino acids 20–914), different fragments of the leucine-rich repeat domain of ZmMSP1 (amino acids are indicated in the Figure) or the empty prey vector (control). Yeast growth was monitored in a dilution series on high stringency (for interaction) and medium stringency media (for growth control).