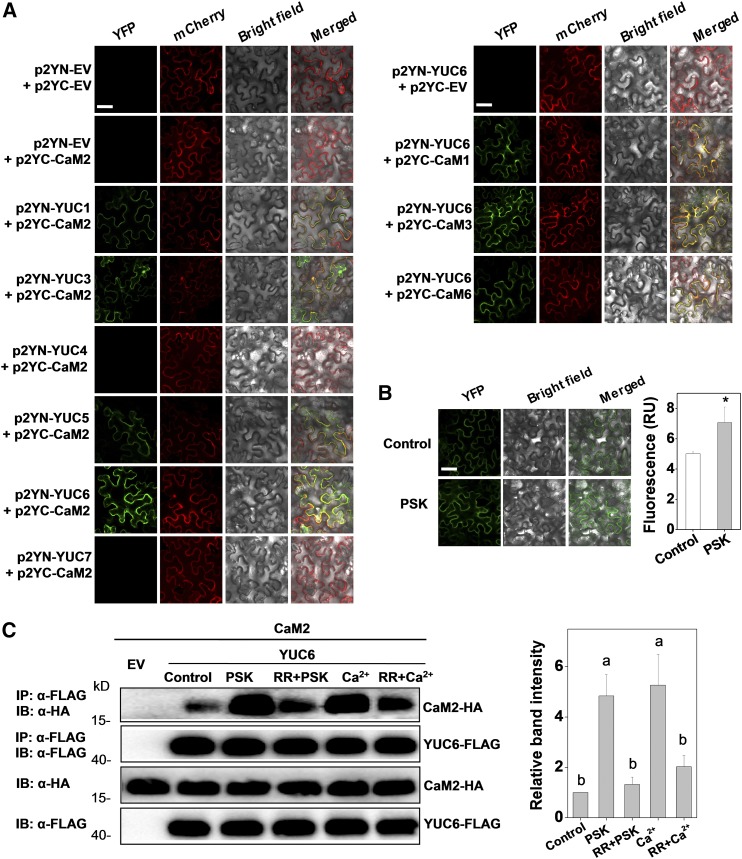
Figure 6.
Tomato CaMs Bind to Auxin Biosynthetic Protein YUCs.
(A) BiFC analyses of the binding between CaM2 and YUCs (left set of panels) and between CaMs and YUC6 (right set of panels). Both spliced YFP constructs and FLS2-mCherry (marker for plasma membrane localization) plasmids were transiently coexpressed in N. benthamiana leaves. The YFP and mCherry signals were visualized under confocal microscopy 48 h after infiltration. Bar = 50 µm.
(B) Changes in the BiFC fluorescence signal between p2YC-CaM2 and p2YN-YUC6 with or without application of PSK (10 µM) for 2 h. Bar = 50 µm. The fluorescence signal intensity from three independent repeats was quantified and the data are shown as means ± sd (n = 3 leaves from different plants). Asterisks indicate a significant effect of PSK application (P < 0.05, Tukey’s test).
(C) Co-IP analysis of the association between HA-tagged CaM2 and FLAG-tagged YUC6 with or without application of 10 µM PSK, 20 µM CaCl2, and 20 μM Ca2+ channel inhibitor RR for 2 h. Total proteins were extracted from leaves transiently expressing the CaM2-HA, YUC6-FLAG construct alone or their combinations after 48 h of infiltration. The extracted proteins were immunoprecipitated with an anti-FLAG antibody and the presence of CaM2-HA and YUC6-FLAG in the immune complex was determined by immunoblot (IB) with the indicated antibody. The co-IP band intensity (top) from three independent repeats was quantified by Image J software. The data are shown as mean ± sd (n = 3 leaves from different plants). Different letters indicate significant differences between treatments (P < 0.05, Tukey’s test).
The experiments in (A) and (B) were repeated three times, and experiments in (C) were repeated two times with similar results.