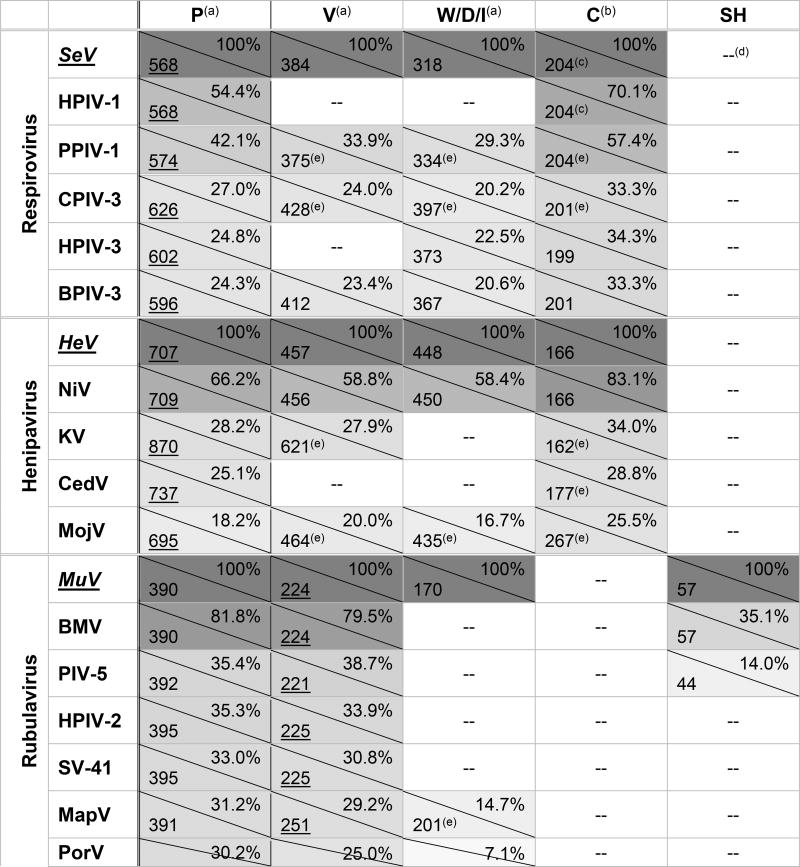
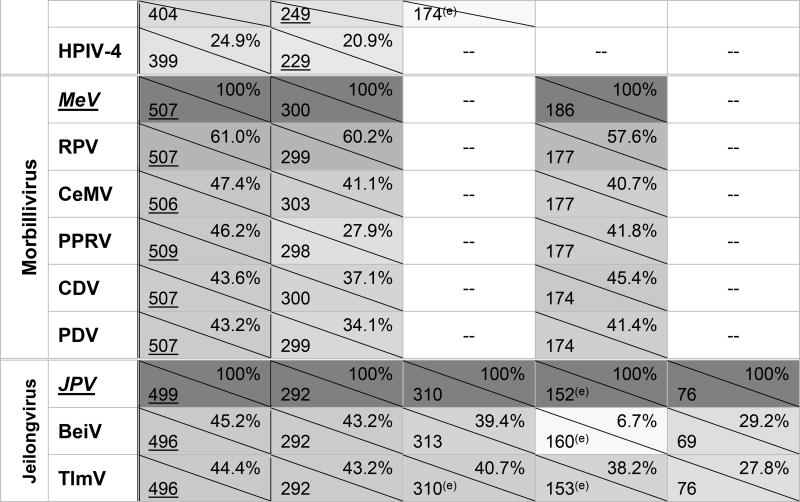
Table 1. Paramyxovirus accessory proteins of interest.
Sizes are given in amino acids (aa; bottom left corner) while percent amino acid identity (progressive pairwise alignment, calculated with default settings of ClustalW in MEGA7.0) to each genus’ type species is given in upper right corner; boxes are shaded to represent percent amino acid identity.
Rubulavirus V proteins (underlined) are the primary transcripts, with P/I proteins being produced through mRNA editing. For all other genera, P proteins (underlined) are the primary transcript, with V/W/D proteins produced through mRNA editing.
C proteins are produced from alternate start codons of the P ORF and are translated in a different open reading frame.
SeV and HPIV-1 produce additional alternate start codon proteins in the same reading frame as C, named C', Y1, and Y2. They share the majority of their sequence with C, but with slightly longer (C') or shorter (Y1, Y2) N termini.
-- = not present or not expressed
putative