ABSTRACT
Shigella is a leading cause of dysentery worldwide, responsible for up to 165 million cases of shigellosis each year. Shigella is also recognised as an exceptional model pathogen to study key issues in cell biology and innate immunity. Several infection models have been useful to explore Shigella biology; however, we still lack information regarding the events taking place during the Shigella infection process in vivo. Here, we discuss a selection of mechanistic insights recently gained from studying Shigella infection of zebrafish (Danio rerio), with a focus on cytoskeleton rearrangements and cellular immunity. We also discuss how infection of zebrafish can be used to investigate new concepts underlying infection control, including emergency granulopoiesis and the use of predatory bacteria to combat antimicrobial resistance. Collectively, these insights illustrate how Shigella infection of zebrafish can provide fundamental advances in our understanding of bacterial pathogenesis and vertebrate host defence. This information should also provide vital clues for the discovery of new therapeutic strategies against infectious disease in humans.
KEY WORDS: Antimicrobial resistance, Autophagy, Cytoskeleton, Emergency granulopoiesis, Inflammation, Macrophage, Neutrophil, Septin, Shigella, Zebrafish
Summary: Here, we review how studying Shigella infection of zebrafish has illuminated novel research avenues in both infection and cell biology.
Introduction
Shigella are a pathovar (see Glossary, Box 1) of Escherichia coli that cause dysentery (see Glossary, Box 1) via inflammatory destruction of the intestinal epithelium, a disease process called shigellosis. Up to 165 million cases of shigellosis are estimated to occur annually, resulting in up to half a million deaths (Lima et al., 2015; Kotloff et al., 2017). Moreover, Shigella infection can give rise to serious postinfectious sequelae (see Glossary, Box 1), such as arthritis, sepsis, seizures and haemolytic uremic syndrome (see Glossary, Box 1). Similar to other Gram-negative (see Glossary, Box 1) pathogens, cases of Shigella with acquired resistance to fluoroquinolones (see Glossary, Box 1) and other antibiotics are rising (Harrington, 2015), and the World Health Organization (WHO) has listed Shigella among its top 12 priority pathogens requiring urgent action (WHO report, 2017). Among the four Shigella subgroups, Shigella flexneri is the most common cause of dysentery in low-income countries and the most prevalent of the Shigella subgroups in children under 5 years of age (Connor et al., 2015). By contrast, infection from Shigella sonnei predominates in developed countries (Kotloff et al., 1999; Holt et al., 2012; Kotloff et al., 2017). The infection process of S. sonnei remains poorly understood compared to that of S. flexneri, and therefore the majority of our current knowledge is extrapolated from work performed using S. flexneri. Important differences between subgroups have been described genetically, but have not been fully tested in infection models, for example the presence of a chromosomally encoded type VI secretion system (T6SS; see Glossary, Box 1) in S. sonnei, which is absent in S. flexneri.
Box 1. Glossary.
Actin tails: Propulsive tails that result from the polymerisation of the host cell actin by intracytosolic pathogens to aid them in disseminating from cell-to-cell.
ASC speck: A platform for caspase-1 activity and readout for inflammasome activation.
Autophagosome: Double-membraned vesicle that compartmentalises cellular material targeted to autophagy.
Autophagy: A highly coordinated process of intracellular degradation whereby cytosolic components are isolated within a double-membrane vacuole (autophagosome) and targeted for lysosomal destruction.
Bdellovibrio bacteriovorus: A Gram-negative bacterium that parasitises other Gram-negative bacteria by invading their periplasmic space, undergoing replication, and killing their prey.
Caspase: Cysteine protease controlling inflammation and programmed cell death.
Cell-autonomous immunity: The ability of a host cell to independently eliminate infectious agents using antimicrobial defences and host cell death.
CRISPR/Cas9: A genome editing approach adapted from the antibacteriophage defence system discovered in bacteria.
Cytokinetic furrow: A micron-scale invagination of the cellular surface during cytokinesis, leading to cell division.
Dysentery: Gastroenteritis resulting in bloody diarrhoea.
E3 ubiquitin ligase: Ligating (E3) enzymes that, together with ubiquitin activating (E1) and conjugating (E2) enzymes, mediate ubiquitylation (a post-translational modification of proteins).
Emergency granulopoiesis: De novo generation of neutrophils that arise from increased myeloid progenitor cell proliferation in response to infection and leukocyte exhaustion.
Haemolytic uremic syndrome: A life-threatening condition caused by the destruction of red blood cells.
Fluoroquinolones: A family of broad spectrum antibacterial agents used in human and veterinary medicine.
Gram-negative bacteria: A group of bacteria that lose the Crystal Violet dye in the Gram's method of staining owing to the structure of their cell wall.
Guanylate-binding proteins: A family of GTPases induced by IFNγ, and key components of cellular immunity.
Haematopoietic stem and progenitor cells: Cells that proliferate (into haematopoietic stem cells) and differentiate (into neutrophils) to mediate emergency granulopoiesis.
Inflammasome: A multi-protein complex and component of the innate immune system that promotes the maturation of inflammatory cytokines through recruitment of Caspase-1.
Interferon regulatory factor 8: A transcription factor required for lineage commitment and myelopoiesis.
Larva: The juvenile form zebrafish undergo before developing into adults.
Macropinocytosis: A nonselective, actin-dependent mechanism of cellular uptake whereby plasma membrane protrusions fold inwards to form vesicles (termed macropinosomes).
Mammalian target of rapamycin: A highly conserved kinase used by cells for nutrient sensing.
Metronidazole: A pro-drug used on transgenic fish (engineered to express nitroreductase using a cell/tissue-specific promoter) to ablate specific cells/tissues.
Morphant: An organism which has been genetically manipulated using morpholino oligonucleotides.
Morpholino oligonucleotide: ∼25 base nucleic acid analogues that affect RNA maturation or translation by sequence-specific base-pairing.
Mycobacterium leprae: A species of bacteria that is the causative agent of leprosy in humans.
Mycobacterium tuberculosis: A species of bacteria that is the causative agent of tuberculosis in humans.
Neural Wiskott-Aldrich syndrome protein: Induces actin polymerisation through the actin-related protein (Arp2/3) complex; it is a specific ligand for Shigella IcsA.
Neutropenia: A condition in which neutrophil number is decreased.
Neutrophil extracellular traps: Net-like structures comprised largely of decondensed chromatin that are released by neutrophils at sites of acute or chronic inflammation.
Nonmuscle myosin II: Actin-binding protein with contractile properties.
Orthologues: Genes in different species that evolved from a common ancestral gene.
Paralogues: Two or more genes that derive from the same ancestral gene, originating via genetic duplication.
Pathovar: Bacterial strains with similar characteristics.
Peptidoglycan: A polymer of amino acids and sugars that comprises the bacterial cell wall.
Phagocytic cup: A micron-scale cup-shaped invagination of the cell membrane formed during phagocytosis.
Phagocytosis: A process by which cells engulf particles, including bacterial pathogens.
Prostaglandin D2: A type of lipid signalling molecule produced at sites of tissue damage or infection to control inflammation.
Pyroptosis: A highly inflammatory form of programmed cell death that can occur in response to the presence of intracellular bacteria.
Salmonella enterica serovar Typhimurium: A zoonotic (transmitted from animals) pathogen that causes gastroenteritis and inflammation of the intestinal mucosa.
Septins: A highly conserved family of GTP binding proteins that interact with the membrane and actin to form higher-order structures including filaments, rings and cages.
Sequelae: Chronic conditions resulting from infection or injury.
Tumour necrosis factor: A monocyte-derived pleiotropic pro-inflammatory cytokine involved in a spectrum of biological processes, such as induction of apoptosis.
Type III secretion system: A membrane embedded needle-like structure present in Gram-negative bacteria used to inject effector proteins into a host cell.
Type VI secretion system: A contractile nanomachine used by Gram-negative bacteria to puncture target cells and deliver effectors.
In addition to being an urgent health threat, S. flexneri is recognised as a paradigm for the investigation of cell biology and innate immunity (Picking and Picking, 2016). Through decades of work performed in vitro using the infection of cultured cells, Shigella has been a valuable model for dissecting how bacteria can invade nonphagocytic cell types (Cossart and Sansonetti, 2004; Haglund and Welch, 2011), form actin tails (see Glossary, Box 1) for cell-to-cell spread (Haglund and Welch, 2011; Welch and Way, 2013), and be recognised by cellular immunity (see Glossary, Box 1) for host defence (Phalipon and Sansonetti, 2007; Ashida et al., 2011, 2015). In an effort to fully decipher the molecular and cellular mechanisms underlying the Shigella infection process, the field is progressively shifting towards in vivo investigation using relevant animal models.
The zebrafish infection model
To date, our capability of understanding the Shigella infection process in vivo has been limited. Although no nonprimate animal model exists that closely mimics shigellosis in humans, a variety of steps underlying the Shigella infection process can be examined using the rabbit (Arm et al., 1965; Perdomo et al., 1994; Schnupf and Sansonetti, 2012), guinea pig (Sereny, 1955; Shim et al., 2007) and mouse models (Yang et al., 2014; Li et al., 2017). Although these mammalian models have provided significant advances in testing mechanisms underlying Shigella pathogenesis, they remain poorly suited for in vivo imaging of the cell biology of Shigella infection.
There are many advantages to using zebrafish larvae (see Glossary, Box 1) to study infection, including their rapid development, fully annotated genome (which is highly homologous to that of humans) and optical accessibility for noninvasive real-time imaging (Lieschke and Currie, 2007; Sullivan et al., 2017). Importantly, zebrafish larvae lack an adaptive immune system during early embryonic development, and thus allow specific study of innate immunity without cross-interference from the adaptive immune system (Lieschke and Trede, 2009). The zebrafish model is also genetically tractable, and therefore amenable to the generation of fluorescent transgenic lines and to targeted gene manipulation. In the case of transient depletion, gene manipulation can be achieved using morpholino oligonucleotides (see Glossary, Box 1; Li et al., 2016). However, morpholinos can elicit off-target effects, and alternative strategies might be required to validate the conclusions. In the case of stable genome editing, mutants can be efficiently generated using zinc finger nucleases (ZFNs) (Foley et al., 2009), transcription activator-like effector nucleases (TALENs) (Bedell et al., 2012), or clustered regulatory interspaced short palindromic repeats/CRISPR-associated protein 9 (CRISPR/Cas9; see Glossary, Box 1; Blum et al., 2015). These systems all induce a site-specific double-stranded break, repaired using a nonhomologous end joining mechanism (Ata et al., 2016). The CRISPR/Cas9 system is currently the most prevalent approach to generate zebrafish mutants.
In recent years, our understanding of the infection process has expanded considerably through the use of the zebrafish infection model (reviewed in Renshaw and Trede, 2012). Originally used to study Mycobacterium marinum, a natural fish pathogen closely related to Mycobacterium tuberculosis (see Glossary, Box 1), zebrafish larvae have since been exploited to study pathogenesis and infection biology using a wide variety of bacteria (Masud et al., 2017; Torraca and Mostowy, 2017), viruses (Levraud et al., 2014; Varela et al., 2017) and fungi (Gratacap and Wheeler, 2014; Yoshida et al., 2017). For this purpose, injection of bacteria in the caudal vein/posterior blood island or Duct of Cuvier has been used to investigate systemic infection responses (Fig. 1), whereas injection in the tail muscle or hindbrain ventricle (HBV) has been used to analyse a directed leukocyte response to a compartmentalised infection (Fig. 1). Zebrafish infection models have been developed to study a variety of enteropathogens. For example, injection of Salmonella enterica serovar Typhimurium (see Glossary, Box 1) into zebrafish has been key for discovery of novel concepts in cellular immunity, immunometabolism, and emergency granulopoiesis (see Glossary, Box 1; reviewed in Torraca and Mostowy, 2017). Recent work has established zebrafish as a model for foodborne enterohaemorrhagic E. coli (EHEC) infection (Stones et al., 2017), a major cause of diarrhoeal illness in humans. Using the protozoan Paramecium caudatum as a vehicle for EHEC delivery, work has shown that zebrafish larvae can be used to study the hallmarks of human EHEC infection, including EHEC-phagocyte interactions in the gut and bacterial transmission to naive hosts (Stones et al., 2017). In the case of Shigella, caudal vein infection of zebrafish was first developed to study Shigella-phagocyte interactions and bacterial autophagy (see Glossary, Box 1) in vivo (Mostowy et al., 2013). Strikingly, many hallmarks of shigellosis observed in humans, including epithelial cell invasion, macrophage cell death and inflammation, are reproduced in a zebrafish model of S. flexneri infection, and are strictly dependent upon the Shigella type III secretion system (T3SS; see Glossary, Box 1). Moreover, studies using this model discovered a scavenger role for neutrophils in eliminating infected macrophages and other cell types that fail to control Shigella infection (Mostowy et al., 2013). In this case, when infected macrophages and other cell types fail to control infection, scavenger neutrophils act as a compensatory mechanism to clear both the dead macrophages and the infection.
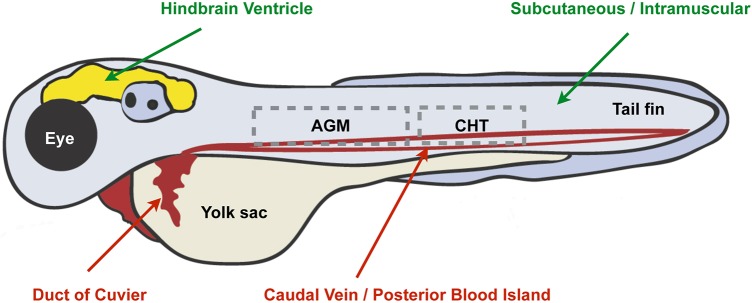
Fig. 1.
Different injection sites of zebrafish larvae used to study Shigella infection. Main attributes of different injection sites used for the study of Shigella infection of zebrafish larvae (3 days postfertilisation). To study systemic infection and Shigella-phagocyte interactions, intravenous injection of Shigella into the circulation is performed via the caudal vein/posterior blood island or Duct of Cuvier (highlighted in red). To study compartmentalised infection and a directed leukocyte response to Shigella, injection of Shigella into the hindbrain ventricle, or subcutaneous/intramuscular injection of Shigella into epithelial cells of the tail muscle, is used (highlighted in green). The dashed line boxes indicate the aorta-gonad-mesonephros (AGM) and caudal hematopoietic tissue (CHT) where emergency granulopoiesis (see Glossary, Box 1) takes place.
Collectively, these reports introduced the zebrafish as a novel animal model to study the cell biology and innate immune response to Shigella at the molecular, cellular and whole-animal levels. In this Review, we highlight the diverse applications of Shigella-zebrafish infection, discussing the progress and insights achieved to date. We also discuss several open questions and future prospects.
Recent mechanistic insights into Shigella infection
Here, we summarise a selection of mechanistic insights recently gained from studying Shigella infection of zebrafish. We focus on examples from two main themes: cytoskeleton rearrangements during infection, a historically important field of study critical for our understanding of host-pathogen interactions, and cellular immunity, a rapidly evolving field important for understanding host defence. These new mechanistic insights significantly expand our knowledge of the host response to Shigella infection, and also shed light on the general mechanisms crucial for host defence against bacterial pathogens.
Cytoskeleton rearrangements during Shigella infection
Investigation of the cytoskeleton during bacterial infection has enabled major discoveries in both infection and cell biology (Cossart and Sansonetti, 2004; Haglund and Welch, 2011; Welch and Way, 2013). For example, how pathogens manipulate the host cytoskeleton to gain entry into cells and polymerise actin tails has revolutionised our understanding of phagocytosis (see Glossary, Box 1) and cell motility (Cossart and Sansonetti, 2004; Haglund and Welch, 2011; Welch and Way, 2013). In this section, we review the host cell response to Shigella invasion and actin-based motility, and discuss what has recently been learned from investigation using zebrafish.
Shigella uptake into nonphagocytic cells has been well characterised in vitro, where entry is dependent upon injection of T3SS effector proteins into the host cell (reviewed in Cossart and Sansonetti, 2004). This form of bacterial uptake, called trigger-mediated entry, causes the reorganisation of the host cell cytoskeleton via actin remodelling and plasma membrane ruffling (Fig. 2A). Engulfment occurs by a process analogous to macropinocytosis (see Glossary, Box 1), and, in the case of Shigella, is followed by vacuolar rupture and escape of bacteria into the cytosol (Weiner et al., 2016). Once in the cytosol, Shigella can recruit neural Wiskott-Aldrich syndrome protein (N-WASp; see Glossary, Box 1) to polymerise actin tails for its own motility (Fig. 2B), a process dependent on the bacterial outer membrane autotransporter IcsA (Goldberg and Theriot, 1995). Studies using human epithelial cells have discovered key roles for septins (see Glossary, Box 1) in the regulation of actin-mediated infection processes (reviewed in Torraca and Mostowy, 2016). A relatively poorly understood component of the cytoskeleton compared to actin, septins are important for a variety of cellular processes including cytokinesis and host-pathogen interactions (reviewed in Saarikangas and Barral, 2011; Mostowy and Cossart, 2012). Septins are highly conserved in vertebrates, and in humans are categorised into four groups (called the SEPT2, SEPT3, SEPT6 and SEPT7 groups), the products of which assemble into hetero-oligomeric complexes, filaments and ring-like structures. Septins can recognise areas of micron-scale curvature, including the cytokinetic furrow (see Glossary, Box 1) and phagocytic cup (see Glossary, Box 1), where they act both as scaffolds for protein recruitment and as diffusion barriers for subcellular compartmentalisation (Saarikangas and Barral, 2011; Mostowy and Cossart, 2012; Bezanilla et al., 2015; Cannon et al., 2017). Consistent with this, new work has shown that septins in neurons contribute to cell shape memory (Boubakar et al., 2017) and the maturation of dendritic spines (Yadav et al., 2017).
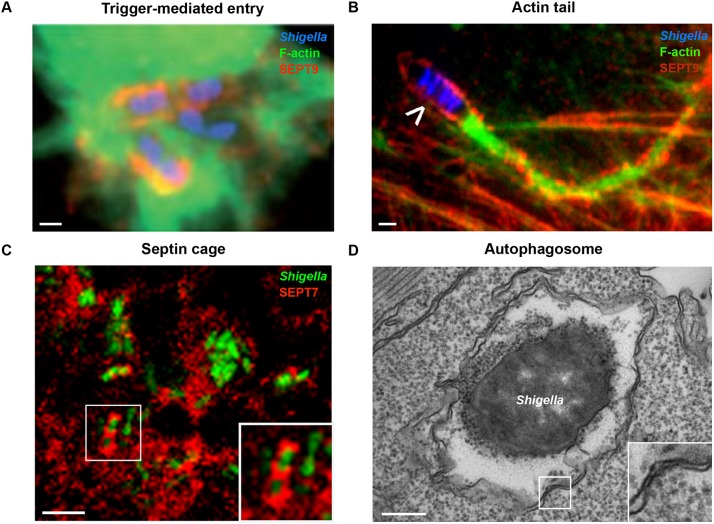
Fig. 2.
The hallmarks of Shigella infection. (A) Trigger-mediated entry by Shigella. HeLa cells were infected with Shigella (blue), fixed for fluorescent microscopy, and labelled with antibodies to SEPT9 (red) and phalloidin for F-actin (green) to highlight septin recruitment at the site of Shigella entry. Scale bar: 1 µm. (B) The Shigella actin tail. HeLa cells were infected with Shigella (blue; white arrowhead indicates a motile bacterium) for 3 h, fixed for fluorescent microscopy, and labelled with antibodies to SEPT2 (red) and phalloidin for F-actin (green) to highlight septin ring formation around the actin tails. Scale bar: 1 µm. (C) The Shigella-septin cage in vivo. SEPT7 (red) assembles into cage-like structures around S. flexneri (green). Zebrafish larvae were infected with green fluorescent protein (GFP)-Shigella for 4 h, fixed, labelled with antibodies to SEPT7 and imaged by confocal microscopy. The inset shows a higher magnification view of the boxed region in C, showing Shigella entrapped within a septin cage. Scale bar: 5 µm. (D) An autophagosome sequestering cytosolic Shigella in vivo. Zebrafish larvae were infected in the tail muscle with GFP-Shigella for 4 h and fixed for electron microscopy. The inset shows a higher magnification view of the boxed region in D, showing the double membrane, a hallmark of autophagosomes. Scale bar: 0.25 µm. Images adapted from Mostowy and Cossart (2009) (A), Mostowy et al. (2010) (B) and Mostowy et al. (2013) (C,D).
During Shigella infection, septins are recruited to the phagocytic cup alongside actin and form rings around invading bacterium (Mostowy et al., 2009a,b). Although the precise role of septins during bacterial entry is not clear, the depletion of SEPT2 by small interfering (si)RNA significantly reduces Shigella entry into host cells (Mostowy et al., 2009a). Following bacterial escape from the phagosome to the cytosol, septins are recruited to actin-polymerising bacteria, forming cage-like structures around Shigella that inhibit cell-cell spread, in a process called septin caging (Mostowy et al., 2010). The depletion of SEPT2, SEPT9 or nonmuscle myosin II (see Glossary, Box 1) inhibits septin caging and increases the number of bacteria with actin tails, whereas increasing SEPT2-nonmuscle myosin II interactions using tumour necrosis factor (TNF; see Glossary, Box 1) increases septin caging and prevents the formation of actin tails (Mostowy et al. 2010). Importantly, septin cages have been observed in zebrafish cells in vitro, as well as in vivo (Fig. 2C), supporting their role as an evolutionarily conserved host defence assembly (Mostowy et al., 2013).
The investigation of septins in vivo using mouse models has been challenging, considering that their deletion can often result in embryonic lethality (reviewed in Kinoshita and Noda, 2001; Mostowy and Cossart, 2012). However, developmental studies using zebrafish have linked the depletion of some septins (i.e. Sept6, Sept9a, Sept9b, Sept15) to growth defects and aberrant left-right asymmetry (Landsverk et al., 2010; Dash et al., 2014, 2016; Zhai et al., 2014). Zebrafish septins have also been shown to play a crucial role during S. flexneri infection (Mazon-Moya et al., 2017). In this case, depletion of Sept15 or Sept7b [zebrafish orthologues (see Glossary, Box 1) of human SEPT7] significantly increases host susceptibility both to compartmentalised (HBV) and systemic (caudal vein) Shigella infection. Live-cell imaging of Sept15-depleted larvae during infection revealed a failure of neutrophils to control infection. Susceptibility of septin morphants (see Glossary, Box 1) to infection is strictly dependent on the Shigella T3SS, as infection with T3SS-deficient S. flexneri is equally controlled by control or Sept15 morphants (Mazon-Moya et al., 2017). Sept15 morphants also exhibit neutropenia (see Glossary, Box 1; Mazon-Moya et al., 2017), a marker of poor prognosis of infection in humans. These data are consistent with experiments using Sept7-deficient transgenic mice showing impaired granulopoietic potential of myeloid progenitors (Menon et al., 2014). The use of morpholinos to target interferon regulatory factor 8 (Irf8; see Glossary, Box 1) and boost neutrophil numbers in Sept15 morphants failed to improve the outcome from Shigella infection, suggesting a cell-autonomous defect of neutrophils with nonfunctional Sept15 (Mazon-Moya et al., 2017). Collectively, studies of zebrafish infection with Shigella illustrate a new role for septins in host defence against bacterial infection, and highlight the crucial role of the cytoskeleton in neutrophil development and behaviour.
Cellular immunity to Shigella infection
Shigella has emerged as a model pathogen to investigate how bacteria can be recognised by, or escape from, the host immune system (reviewed in Phalipon and Sansonetti, 2007; Ashida et al., 2011, 2015). For example, studies using Shigella have helped to discover major roles for nucleotide-binding oligomerisation domain (NOD)-like receptors (NLRs) (Girardin et al., 2003), bacterial autophagy (Ogawa et al., 2005) and inflammasomes (see Glossary, Box 1; Willingham et al., 2007) in host defence. This section will discuss cellular immunity to Shigella, and how this knowledge has evolved following recent in vivo studies using zebrafish.
Shigella-autophagy interactions
Discovered as a nutrient recycling process in response to starvation (reviewed in Yang and Klionsky, 2010), autophagy is also a selective process crucial for cellular immunity (reviewed in Levine et al., 2011). During Shigella infection, cytosolic bacteria are recognised by the autophagy machinery and targeted for lysosomal delivery (Ogawa et al., 2005). The cytoskeleton, well known for its role in bacterial invasion and motility, also plays an important role during the autophagy response to bacteria (reviewed in Mostowy, 2014; Mostowy and Shenoy, 2015). Work using human epithelial cells has shown that septin cages colocalise with ubiquitylated proteins that concentrate around autophagy-targeted Shigella (Mostowy et al., 2010). Consistent with the notion that septin cage assembly and autophagy of bacteria are interdependent, the number of Shigella septin cages is significantly reduced following siRNA knockdown of autophagy machinery components (Mostowy et al., 2010; Mostowy et al., 2011). Moreover, Shigella mutants lacking IcsB, a T3SS effector involved in autophagy evasion, are compartmentalised into septin cages more efficiently than their wild-type counterparts (Mostowy et al., 2010; Mostowy et al., 2011). Importantly, ∼50% of Shigella entrapped in septin cages are metabolically inactive, highlighting septin caging as an antibacterial mechanism (Sirianni et al., 2016). In one of the first studies to use zebrafish for investigation of bacterial autophagy in vivo (Mostowy et al., 2013), infection of larvae with S. flexneri revealed that autophagy is a crucial component of innate immunity at the whole-organism level (Fig. 2D). In this case, depletion of the autophagy receptor protein p62 significantly reduced septin cage-associated S. flexneri, and increased bacterial burden and host mortality following infection. How septin cages assemble and interact with other cytoskeletal proteins during the autophagy response to bacteria, and their ability to be manipulated for therapy, is currently the focus of intense investigation.
Following T3SS-mediated invasion of epithelial cells, a number of host cell mechanisms recruit the autophagy machinery to Shigella, including the NLRs NOD1 and NOD2 that detect bacterial peptidoglycan (see Glossary, Box 1) in the cytosol (reviewed in Krokowski and Mostowy, 2016; Keestra-Gounder and Tsolis, 2017). NOD1/2 receptors can recruit ATG16L1, a protein crucial for autophagosome (see Glossary, Box 1) development, and inhibit the release of proinflammatory cytokines (Sorbara et al., 2013). A separate autophagy pathway follows phagocytic rupture, in which the autophagy machinery is recruited to damaged phagosomal membranes labelled by ubiquitin (Dupont et al., 2009; Thurston et al., 2012). In the cytosol, ATG5 interacts with the bacterial IcsA and targets Shigella towards autophagy and septin caging (Ogawa et al., 2005; Mostowy et al., 2010). A role for mitochondria in the promotion of septin cage assembly and bacterial autophagy has also been demonstrated (Sirianni et al., 2016). To avoid autophagic destruction, actin-polymerising S. flexneri can fragment mitochondria to counteract the assembly of septin cages (Sirianni et al., 2016). Rapamycin, a pharmacological inhibitor of mammalian target of rapamycin (mTOR; see Glossary, Box 1), stimulates autophagy and has been proposed as a therapy to promote bacterial clearance (Abdel-Nour et al., 2014). Indeed, in vitro experimental work in a variety of cultured cell models has shown that rapamycin can enhance the killing of bacteria (Gutierrez et al., 2004; Tattoli et al., 2012). However, rapamycin treatment of S. flexneri-infected zebrafish failed to rescue the infected hosts (Mostowy et al., 2013), suggesting that therapeutic manipulation of autophagy in vivo is complex. Similar results were observed following rapamycin treatment of M. marinum-infected zebrafish, which also failed to rescue the infected hosts (van der Vaart et al., 2014).
Shigella-inflammasome interactions
In human (Zychlinsky et al., 1992) and zebrafish (Mostowy et al., 2013) macrophages, Shigella infection induces cell death, yet the underlying mechanisms are not fully understood. Interestingly, targeted ablation of nascent macrophages in transgenic zebrafish larvae Tg(mpeg1:Gal4-FF)gl25/Tg(UASE1b:nfsB.mCherry)c26 using metronidazole (see Glossary, Box 1) decreased larval survival following S. flexneri infection (Mazon-Moya et al., 2017), indicating that macrophages can somehow protect against infection in vivo. Experimental work performed in vitro using cultured cell models has shown that Shigella can manipulate inflammasome activity (reviewed in Hermansson et al., 2016; Suzuki et al., 2017). Inflammasome-triggered cleavage of interleukin (IL)-1β and IL-18 via cysteine-aspartic protease (caspase)-1 (see Glossary, Box 1) recruitment results in pyroptosis (see Glossary, Box 1; Miao et al., 2011). A variety of exciting papers have used zebrafish to study inflammasome activation in vivo. For example, clearance of S. Typhimurium from zebrafish by neutrophils requires the interferon gamma (IFNγ)-inducible GTPase guanylate-binding protein-4 (GBP-4; see Glossary, Box 1) and the inflammasome-dependent production of prostaglandin D2 (see Glossary, Box 1; Tyrkalska et al., 2016). In a separate study, zebrafish infection with a strain of Listeria monocytogenes ectopically expressing flagellin, the principal component of bacterial flagella, was shown to activate the inflammasome in macrophages and decrease infection (Vincent et al., 2016). Although tools to study the inflammasome in zebrafish remain limited compared to mice, zebrafish can now be used to follow the assembly of apoptosis-associated speck-like protein containing a caspase-recruitment domain (ASC) specks (see Glossary, Box 1) in vivo (Kuri et al., 2017). It will next be of great interest to study the events and pathways underlying inflammasome activation and cell death during Shigella infection of zebrafish.
The host cytoskeletal components drive activation of the inflammasome and cell-autonomous immunity (see Glossary, Box 1; reviewed in Mostowy and Shenoy, 2015). Work has linked actin to inflammation control through regulation of the NACHT, LRR and PYD domain-containing protein 3 (NLRP3) (Pelegrin and Surprenant, 2009; Jin et al., 2013; Johnson et al., 2013) and pyrin inflammasome complexes (Kim et al., 2015; Standing et al., 2017). Actin polymerisation proteins can regulate the NLRP3 inflammasome by interacting with caspase-1 and other inflammasome components (Pelegrin and Surprenant, 2009; Jin et al., 2013; Johnson et al., 2013). Actin depolymerisation, as a consequence of mutations in WD repeat-containing protein (WDR1), can activate the pyrin inflammasome (Kim et al., 2015; Standing et al., 2017). In the case of S. flexneri infection, disruption of WASp results in increased inflammasome activity and decreased bacterial clearance (Lee et al., 2017). Recent work using the Shigella-zebrafish infection model has discovered a new role for septins in inflammation control. Experiments in this study showed that Sept15 morphants exhibit significantly increased caspase-1 activity and cell death, indicating that septins restrict inflammation in vivo (Mazon-Moya et al., 2017). Considering that increased inflammasome activity results in increased levels of IL-1β (reviewed in Malik and Kanneganti, 2017), blocking IL-1β signalling represents an attractive therapeutic solution to reduce inflammation. Indeed, inflammation in Shigella-infected Sept15 zebrafish morphants can be dampened by treatment with anakinra, an IL-1 receptor antagonist used to treat inflammatory disorders in humans, which ultimately rescues neutrophil and host survival (Mazon-Moya et al., 2017).
Together, these examples illustrate that zebrafish larvae are well suited to study the breadth of cellular immune responses available to control Shigella infection and inflammation. Moving forward, it can be expected that studies using zebrafish will continue to identify the molecular determinants and events underlying cellular immunity to Shigella infection in vivo.
New ways to control Shigella infection
The Shigella-zebrafish infection model is highly versatile, and can be used to illuminate novel research aiming to control bacterial infection. In this section, we discuss how infection of zebrafish can be used to investigate new concepts underlying infection control, including emergency granulopoiesis and the use of predatory bacteria to combat antimicrobial resistance.
Emergency granulopoiesis
How infection and inflammation can mediate emergency granulopoiesis is a subject that is difficult to fully investigate in vitro. The zebrafish model, however, has recently provided valuable insights into the fundamental aspects of stem cell biology and host defence. Haematopoiesis is the hierarchical process by which haematopoietic stem and progenitor cells (HSPCs; see Glossary, Box 1) produce mature blood cells (King and Goodell, 2011; Jagannathan-Bogdan and Zon, 2013; Laurenti and Göttgens, 2018). Following bacterial infection, circulating populations of neutrophils can become exhausted. Although emergency granulopoiesis can replenish the exhausted neutrophils, the underlying molecular events are mostly unknown (Manz and Boettcher, 2014; Teng et al., 2017). In the zebrafish, emergency granulopoiesis takes place in the aorta-gonad-mesonephros (AGM) and caudal hematopoietic tissue (CHT), and is supplied by the proliferation and differentiation of HSPCs (Fig. 1). Strikingly, a zebrafish model of S. Typhimurium infection revealed a direct expansion of the HSPC niche in response to bacterial infection (Hall et al., 2012). The HSPC expansion arises by release of granulopoietic cytokines, including granulocyte colony-stimulating factor (GCSF; CSF3) (Stachura et al., 2013). Neutrophil expansion in the AGM and CHT is dependent on inducible nitric oxide synthase (iNOS) and the transcription factor CCAAT/enhancer-binding protein β (C/EBPβ) (Hall et al., 2012). Considering that Shigella induces inflammation and neutropenia during infection of zebrafish (Mazon-Moya et al., 2017), it will now be of great interest to use Shigella infection of zebrafish to further study HSPC signalling and emergency granulopoiesis, and their precise role in host defence. An in-depth understanding of the cellular and molecular mechanisms governing HSPC biology will be important to inform strategies for using the therapeutic manipulation of emergency granulopoiesis to treat bacterial infections in humans. As a natural programme of host defence, strategies to boost emergency granulopoiesis have great therapeutic potential; however, such treatments must balance immune stimulation with the risk of stem cell exhaustion.
Predatory bacteria work alongside the host immune cells to treat Shigella infection
The emergence of antimicrobial resistant (AMR) bacteria is a global health threat, and improved antimicrobial therapies are urgently needed. The ESKAPE pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa and Enterobacter species) are considered priority pathogens most likely to escape bactericidal treatment (Boucher et al., 2009), and are also recognised as new paradigms in microbial pathogenesis, transmission and resistance (Pendleton et al., 2013). An increased understanding of these pathogens can therefore lead to innovative strategies in the fight against antibiotic-resistant infections. Bdellovibrio bacteriovorus (see Glossary, Box 1) is a species of predatory bacteria gaining recognition for its ability to invade and kill Gram-negative pathogens (reviewed in Tyson and Sockett, 2017). Bdellovibrio have great potential to be used as ‘living antibiotics’ because they target pathogens using mechanisms that make acquiring resistance to predation difficult. Work has shown that Bdellovibrio can predate upon a wide variety of AMR Gram-negative bacteria in vitro (Kadouri et al., 2013). However, despite our understanding of bacterial predation in vitro, not much is known about the efficacy of bacterial predation in vivo. A recent study examined Bdellovibrio treatment following intranasal inoculation of rats with sublethal doses of K. pneumoniae (Shatzkes et al., 2016). In this case, the authors found a 2-log reduction in Klebsiella burden in the lungs of the Bdellovibrio-treated rats. However, a follow-up study revealed that exposure of Klebsiella-infected rats to successive rounds of Bdellovibrio is ineffective against an acute blood infection (Shatzkes et al., 2017). One caveat of using mammalian models is that it is difficult to visualise the predator-prey interactions in vivo. To circumvent this, zebrafish larvae infected with multidrug resistant Shigella were used to study Bdellovibrio predation in vivo (Willis et al., 2016). This model system enabled, for the first time, the visualisation of bacterial predator-prey interactions in vivo in the presence or absence of an innate immune system (Fig. 3). In this study, immunocompromised larvae were generated using a morpholino targeting the transcription factor Pu.1 (Spi1b), which prevents myeloid cell differentiation. Control and Pu.1 morphants were injected with a lethal dose of Shigella in the HBV and treated with either phosphate buffered saline (PBS) or Bdellovibrio (Willis et al., 2016). Enumerations of Shigella from larval homogenates showed that treatment with Bdellovibrio reduced Shigella numbers in both immunocompromised and immunocompetent zebrafish larvae, but survival was significantly greater in the immunocompetent larvae. These experiments revealed that Bdellovibrio can work in synergy with the host immune cells (macrophages and neutrophils) to clear Shigella infection in vivo, before being cleared by the zebrafish immune system themselves. Next, an in-depth investigation of Bdellovibrio predation on a range of Gram-negative pathogens using the zebrafish model is needed before ultimately translating this procedure to higher vertebrates and humans.
Fig. 3.
Interaction of the predatory bacteria Bdellovibrio with Shigella in the zebrafish. (A) Wild-type zebrafish larvae were injected in the hindbrain ventricle (HBV) at 3 days postfertilisation with >5×103 colony forming units (CFUs) of GFP-S. flexneri (green), followed by hindbrain injection of either PBS or 1-2×105 plaque forming units (PFUs) of mCherry-Bdellovibrio (red), 30-90 min after the initial Shigella infection. Representative images of the HBV in PBS- or Bdellovibrio-treated zebrafish larvae infected with Shigella are shown. The dashed line box shows the region of interaction between fluorescent Bdellovibrio and Shigella. For both treatments, the same larva was imaged over time. Scale bar: 100 µm. hpi, hours postinfection. (B) Representative images of Shigella predation by Bdellovibrio in vivo imaged by high-resolution confocal microscopy. Frames captured over time show stages of Bdellovibrio (red) invasive predation and rounding of Shigella (green). Scale bar: 2.5 µm. mpi, minutes postinfection. Adapted from Willis et al. (2016).
Conclusions and emerging perspectives
Here, we have illustrated how the zebrafish model can be used to study a variety of mechanisms underlying the host response to Shigella infection. The Shigella-zebrafish infection model is innovative because it is applicable to the in vivo milieu and generalisable across infectious agents, thus facilitating the comparison of evolutionarily distinct pathogens. In agreement with this, lessons learned from Shigella infection of zebrafish can be applied to other human bacterial pathogens (e.g. Listeria, mycobacteria, Salmonella), and also to neglected pathogens (e.g. Burkholderia, Rickettsia, Waddlia), of which little is known. Furthermore, Shigella infection of zebrafish can reveal general mechanisms of host defence relevant to combatting human infection from viral and fungal pathogens.
As an emerging animal model for the study of Shigella infection, the zebrafish offers numerous advantages and an exciting array of cell biology applications. However, there are some limitations to using the zebrafish model to study human bacterial infection. For example, there is a demand for improved tools, such as zebrafish-specific antibodies and knockout lines generated by CRISPR/Cas9. Another potential caveat is temperature: the optimal zebrafish growth temperature of 28°C mimics their natural environment, and deviations from this can affect their developmental cycle, whereas human pathogens are sometimes more virulent at 37°C, a temperature that in zebrafish can cause a depressed immune function and an upregulation of stress response proteins not present at 28°C (Long et al., 2012; Lam et al., 2013). Although zebrafish larvae offer the opportunity to study innate immunity in isolation, they cannot enable studies involving adaptive immunity. Experiments involving adaptive immunity will require the use of adult zebrafish, which have been used to study a variety of pathogenic bacteria, including M. marinum (Oehlers et al., 2015; Cronan et al., 2016) and Mycobacterium leprae (see Glossary, Box 1; Madigan et al., 2017). Lastly, the study of gene function in zebrafish can be challenging owing to the presence of paralogues (see Glossary, Box 1) within the genome (Howe et al., 2013). However, paralogues arising from duplication events in the genome can offer a redundancy, which can be beneficial for studies of genes for which knockout might otherwise result in embryonic lethality. Redundancy in the zebrafish genome has been beneficial to study a variety of genes crucial for host response to infection, including toll-like receptors (TLRs) (reviewed in Kanwal et al., 2014), NLRs (reviewed in Howe et al., 2016), and two Cxcl12 paralogues functioning as chemokines to activate phagocytes (Boldajipour et al., 2008).
As the Shigella-zebrafish model gains traction for the investigation of infection and cellular microbiology in vivo, a number of inspiring future directions are emerging (Box 2). Using this model, it will be of great interest to perform global gene expression profiling during infection for the discovery of new mechanisms underlying host defence, and to screen pharmacological compounds for therapeutic advance. To illuminate host-pathogen interactions, several interesting transcriptomic (Stockhammer et al., 2009; Benard et al., 2016) and proteomic (Díaz-Pascual et al., 2017) profiling studies have been used in zebrafish infected with bacterial pathogens; similar approaches can be used in zebrafish infected with Shigella. Moreover, ‘omics’ of isolated cells (e.g. macrophages or neutrophils) infected with Shigella can be illuminating to decipher bacterial virulence factors and host defence mechanisms important during this interaction. Although Shigella infection of adult zebrafish has not yet been tested, we propose that adult zebrafish could be used to study the adaptive immune response to Shigella, which is crucial to understanding the crosstalk between Shigella and T lymphocytes (reviewed in Salgado-Pabón et al., 2014) or to developing vaccine strategies (reviewed in Phalipon et al., 2008; Mani et al., 2016).
Box 2. Outstanding questions in the field:
Shigella infection of zebrafish is valuable to discover novel mechanisms of cellular immunity. How generalisable are the lessons learned from Shigella infection of zebrafish to other bacterial pathogens?
Can studies in zebrafish help to reveal where Shigella resides in natura outside the human host? Can zebrafish be humanised to emulate a more natural route of Shigella infection?
What is the precise role of the cytoskeleton rearrangements (such as actin tails and septin caging) during Shigella infection in vivo? Can we manipulate the cytoskeleton during infection for a therapeutic benefit?
Big data approaches (i.e. RNA sequencing, bacterial mutant libraries) are useful to discover new host pathways and/or bacterial effectors underlying infection. What can these approaches reveal when applied to Shigella infection of zebrafish?
Can the Shigella-zebrafish infection experiments be used to inform experiments using higher vertebrate animal models and vice versa?
Recent work using mixed bacterial cultures in vitro and in vivo using rabbits and mice has shown that S. sonnei use a T6SS to outcompete S. flexneri (Anderson et al., 2017). These results highlight the T6SS as a key factor underlying the increasing prevalence of S. sonnei in developing countries (Lima et al., 2015; Kotloff et al., 2017), and will likely inspire the use of S. sonnei for cellular microbiology studies using zebrafish. Originally discovered in in vitro studies using Shigella infection of neutrophils, the release of neutrophil extracellular traps (NETs; see Glossary, Box 1) is currently regarded as an important cellular immunity mechanism for controlling a variety of pathogens, including Staphylococcus, Klebsiella, Aspergillus and Candida (Brinkmann et al., 2004; Branzk et al., 2014; Storisteanu et al., 2017). It is tempting to speculate that zebrafish infection models will be valuable to fully dissect the role of NETs in host defence in vivo. Recent work has shown in human cells (Wandel et al., 2017) and mice (Li et al., 2017) that Shigella evades GBPs by releasing IpaH9.8, an E3 ubiquitin ligase (see Glossary, Box 1) that triggers GBP degradation. Shigella infection of zebrafish can therefore be used to study GBPs and their precise role in host defence in vivo.
The field of pathogenesis is evolving from focusing on the mechanics of infection in vitro using cultured cells towards studying the infection process in vivo using animal models. The crypt culture model, known as intestinal organoids, is also powerful to study host-pathogen interactions (Nigro et al., 2016) and the response of the epithelium to enteric pathogens (Nigro et al., 2014). Considering the versatility of zebrafish infection models, it can be predicted that our understanding of Shigella pathogenesis will greatly benefit from in vivo applications of genome editing (e.g. CRISPR/Cas9), high-resolution imaging techniques (e.g. high-resolution fluorescence microscopy and cryo-electron tomography), and automated high-throughput screening approaches. Finally, it will be crucial that advancements in infection and cell biology learned from Shigella infection of zebrafish be applied for further study to higher vertebrates, including humans, and also as a therapeutic avenue in both human and veterinary medicine.
Acknowledgements
We thank Alex Willis and Vincenzo Torraca for commenting on the manuscript.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
This work was supported by the Wellcome Trust (206444/Z/17/Z and WT097411MA) and Lister Institute of Preventive Medicine.
References
- Abdel-Nour M., Tsalikis J., Kleinman D. and Girardin S. E. (2014). The emerging role of mTOR signalling in antibacterial immunity. Immunol. Cell Biol. 92, 346-353. 10.1038/icb.2014.3 [DOI] [PubMed] [Google Scholar]
- Anderson M. C., Vonaesch P., Saffarian A., Marteyn B. S. and Sansonetti P. J. (2017). Shigella sonnei encodes a functional T6SS used for interbacterial competition and niche occupancy. Cell Host Microbe 21, 769-776.e3. 10.1016/j.chom.2017.05.004 [DOI] [PubMed] [Google Scholar]
- Arm H. G., Floyd T. M., Faber J. E. and Hayes J. R. (1965). Use of ligated segments of rabbit small intestine in experimental shigellosis. J. Bacteriol. 89, 803-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashida H., Ogawa M., Mimuro H., Kobayashi T., Sanada T. and Sasakawa C. (2011). Shigella are versatile mucosal pathogens that circumvent the host innate immune system. Curr. Opin. Immunol. 23, 448-455. 10.1016/j.coi.2011.06.001 [DOI] [PubMed] [Google Scholar]
- Ashida H., Mimuro H. and Sasakawa C. (2015). Shigella manipulates host immune responses by delivering effector proteins with specific roles. Front. Immunol. 6, 219 10.3389/fimmu.2015.00219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ata H., Clark K. J. and Ekker S. C. (2016). The zebrafish genome editing toolkit. Methods. Cell Biol. 135, 149-170. 10.1016/bs.mcb.2016.04.023 [DOI] [PubMed] [Google Scholar]
- Bedell V. M., Wang Y., Campbell J. M., Poshusta T. L., Starker C. G., Krug R. G. II, Tan W., Penheiter S. G., Ma A. C., Leung A. Y. H. et al. (2012). In vivo genome editing using a high-efficiency TALEN system. Nature 491, 114-118. 10.1038/nature11537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benard E. L., Rougeot J., Racz P. I., Spaink H. P. and Meijer A. H. (2016). Transcriptomic approaches in the zebrafish model for tuberculosis-insights into host- and pathogen-specific determinants of the innate immune response. Adv. Genet. 95, 217-251. 10.1016/bs.adgen.2016.04.004 [DOI] [PubMed] [Google Scholar]
- Bezanilla M., Gladfelter A. S., Kovar D. R. and Lee W.-L. (2015). Cytoskeletal dynamics: a view from the membrane. J. Cell Biol. 209, 329-337. 10.1083/jcb.201502062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum M., De Robertis E. M., Wallingford J. B. and Niehrs C. (2015). Morpholinos: antisense and sensibility. Dev. Cell 35, 145-149. 10.1016/j.devcel.2015.09.017 [DOI] [PubMed] [Google Scholar]
- Boldajipour B., Mahabaleshwar H., Kardash E., Reichman-Fried M., Blaser H., Minina S., Wilson D., Xu Q. and Raz E. (2008). Control of chemokine-guided cell migration by ligand sequestration. Cell 132, 463-473. 10.1016/j.cell.2007.12.034 [DOI] [PubMed] [Google Scholar]
- Boubakar L., Falk J., Ducuing H., Thoinet K., Reynaud F., Derrington E. and Castellani V. (2017). Molecular memory of morphologies by septins during neuron generation allows early polarity inheritance. Neuron 95, 834-851.e5. 10.1016/j.neuron.2017.07.027 [DOI] [PubMed] [Google Scholar]
- Boucher H. W., Talbot G. H., Bradley J. S., Edwards J. E., Gilbert D., Rice L. B., Scheld M., Spellberg B. and Bartlett J. (2009). Bad bugs, no drugs: no ESKAPE! An update from the infectious diseases society of America. Clin. Infect. Dis. 48, 1-12. 10.1086/595011 [DOI] [PubMed] [Google Scholar]
- Branzk N., Lubojemska A., Hardison S. E., Wang Q., Gutierrez M. G., Brown G. D. and Papayannopoulos V. (2014). Neutrophils sense microbe size and selectively release neutrophil extracellular traps in response to large pathogens. Nat. Immunol. 15, 1017-1025. 10.1038/ni.2987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkmann V., Reichard U., Goosmann C., Fauler B., Uhlemann Y., Weiss D. S., Weinrauch Y. and Zychlinsky A. (2004). Neutrophil extracellular traps kill bacteria. Science 303, 1532-1535. 10.1126/science.1092385 [DOI] [PubMed] [Google Scholar]
- Cannon K. S., Woods B. L. and Gladfelter A. S. (2017). The unsolved problem of how cells sense micron-scale curvature. Trends Biochem. Sci. 42, 961-976. 10.1016/j.tibs.2017.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor T. R., Barker C. R., Baker K. S., Weill F.-X., Talukder K. A., Smith A. M., Baker S., Gouali M., Pham Thanh D., Jahan Azmi I. et al. (2015). Species-wide whole genome sequencing reveals historical global spread and recent local persistence in Shigella flexneri . eLife 4, e07335 10.7554/eLife.07335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossart P. and Sansonetti P. J. (2004). Bacterial invasion: the paradigms of enteroinvasive pathogens. Science 304, 242-248. 10.1126/science.1090124 [DOI] [PubMed] [Google Scholar]
- Cronan M. R., Beerman R. W., Rosenberg A. F., Saelens J. W., Johnson M. G., Oehlers S. H., Sisk D. M., Jurcic Smith K. L., Medvitz N. A., Miller S. E. et al. (2016). Macrophage epithelial reprogramming underlies mycobacterial granuloma formation and promotes infection. Immunity 45, 861-876. 10.1016/j.immuni.2016.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dash S. N., Lehtonen E., Wasik A. A., Schepis A., Paavola J., Panula P., Nelson W. J. and Lehtonen S. (2014). sept7b is essential for pronephric function and development of left–right asymmetry in zebrafish embryogenesis. J. Cell Sci. 127, 1476-1486. 10.1242/jcs.138495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dash S. N., Hakonen E., Ustinov J., Otonkoski T., Andersson O. and Lehtonen S. (2016). sept7b is required for the differentiation of pancreatic endocrine progenitors. Sci. Rep. 6, 24992 10.1038/srep24992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-Pascual F., Ortíz-Severín J., Varas M. A., Allende M. L. and Chávez F. P. (2017). In vivo host-pathogen interaction as revealed by global proteomic profiling of zebrafish larvae. Front. Cell Infect. Microbiol. 7, 334 10.3389/fcimb.2017.00334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont N., Lacas-Gervais S., Bertout J., Paz I., Freche B., Van Nhieu G. T., van der Goot F. G., Sansonetti P. J. and Lafont F. (2009). Shigella phagocytic vacuolar membrane remnants participate in the cellular response to pathogen invasion and are regulated by autophagy. Cell Host Microbe 6, 137-149. 10.1016/j.chom.2009.07.005 [DOI] [PubMed] [Google Scholar]
- Foley J. E., Maeder M. L., Pearlberg J., Joung J. K., Peterson R. T. and Yeh J.-R. J. (2009). Targeted mutagenesis in zebrafish using customized zinc-finger nucleases. Nat. Protoc. 4, 1855 10.1038/nprot.2009.209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girardin S. E., Boneca I. G., Carneiro L. A., Antignac A., Jéhanno M., Viala J., Tedin K., Taha M. K., Labigne A., Zähringer U. et al. (2003). Nod1 detects a unique muropeptide from Gram-negative bacterial peptidoglycan. Science 300, 1584-1587. 10.1126/science.1084677 [DOI] [PubMed] [Google Scholar]
- Goldberg M. B. and Theriot J. A. (1995). Shigella flexneri surface protein IcsA is sufficient to direct actin-based motility. Proc. Natl. Acad. Sci. USA 92, 6572-6576. 10.1073/pnas.92.14.6572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratacap R. L. and Wheeler R. T. (2014). Utilization of zebrafish for intravital study of eukaryotic pathogen–host interactions. Dev. Comp. Immunol. 46, 108-115. 10.1016/j.dci.2014.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez M. G., Master S. S., Singh S. B., Taylor G. A., Colombo M. I. and Deretic V. (2004). Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell 119, 753-766. 10.1016/j.cell.2004.11.038 [DOI] [PubMed] [Google Scholar]
- Haglund C. M. and Welch M. D. (2011). Pathogens and polymers: microbe–host interactions illuminate the cytoskeleton. J. Cell Biol. 195, 7-17. 10.1083/jcb.201103148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall C. J., Flores M. V., Oehlers S. H., Sanderson L. E., Lam E. Y., Crosier K. E. and Crosier P. S. (2012). Infection-responsive expansion of the hematopoietic stem and progenitor cell compartment in zebrafish is dependent upon inducible nitric oxide. Cell Stem Cell 10, 198-209. 10.1016/j.stem.2012.01.007 [DOI] [PubMed] [Google Scholar]
- Harrington R. (2015). Drug-resistant stomach bug. Sci. Am. 313, 88 10.1038/scientificamerican0815-88 [DOI] [PubMed] [Google Scholar]
- Hermansson A. K., Paciello I. and Bernardini M. L. (2016). The orchestra and its maestro: Shigella's fine-tuning of the inflammasome platforms. Curr. Top. Microbiol. Immunol. 397, 91-115. 10.1007/978-3-319-41171-2_5 [DOI] [PubMed] [Google Scholar]
- Holt K. E., Baker S., Weill F.-X., Holmes E. C., Kitchen A., Yu J., Sangal V., Brown D. J., Coia J. E., Kim D. W. et al. (2012). Shigella sonnei genome sequencing and phylogenetic analysis indicate recent global dissemination from Europe. Nat. Genet. 44, 1056-1059. 10.1038/ng.2369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe K., Clark M. D., Torroja C. F., Torrance J., Berthelot C., Muffato M., Collins J. E., Humphray S., McLaren K., Matthews L. et al. (2013). The zebrafish reference genome sequence and its relationship to the human genome. Nature 496, 498-503. 10.1038/nature12111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe K., Schiffer P. H., Zielinski J., Wiehe T., Laird G. K., Marioni J. C., Soylemez O., Kondrashov F. and Leptin M. (2016). Structure and evolutionary history of a large family of NLR proteins in the zebrafish. Open Biol. 6, 160009 10.1098/rsob.160009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagannathan-Bogdan M. and Zon L. I. (2013). Hematopoiesis. Development 140, 2463-2467. 10.1242/dev.083147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J., Yu Q., Han C. Hu X., Xu S., Wang Q., Wang J., Li N. and Cao X. (2013). LRRFIP2 negatively regulates NLRP3 inflammasome activation in macrophages by promoting Flightless-I-mediated caspase-1 inhibition. Nat. Commun. 4, 2075 10.1038/ncomms3075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K. E., Chikoti L. and Chandran B. (2013). Herpes simplex virus 1 infection induces activation and subsequent inhibition of the IFI16 and NLRP3 inflammasomes. J. Virol. 87, 5005-5018. 10.1128/JVI.00082-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadouri D. E., To K., Shanks R. M. Q. and Doi Y. (2013). Predatory bacteria: a potential ally against multidrug-resistant Gram-negative pathogens. PLoS ONE 8, e63397 10.1371/journal.pone.0063397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanwal Z., Wiegertjes G. F., Veneman W. J., Meijer A. H. and Spaink H. P. (2014). Comparative studies of Toll-like receptor signalling using zebrafish. Dev. Comp. Immunol. 46, 35-52. 10.1016/j.dci.2014.02.003 [DOI] [PubMed] [Google Scholar]
- Keestra-Gounder A. M. and Tsolis R. M. (2017). NOD1 and NOD2: beyond peptidoglycan sensing. Trends Immunol. 38, 758-767. 10.1016/j.it.2017.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M. L., Chae J. J., Park Y. H., De Nardo D., Stirzaker R. A., Ko H.-J., Tye H., Cengia L., DiRago L., Metcalf D. et al. (2015). Aberrant actin depolymerisation triggers the pyrin inflammasome and autoinflammatory disease that is dependent on IL-18, not IL-1β. J. Exp. Med. 212, 927-938. 10.1084/jem.20142384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King K. Y. and Goodell M. A. (2011). Inflammatory modulation of HSCs: viewing the HSC as a foundation for the immune response. Nat. Rev. Immunol. 11, 685-692. 10.1038/nri3062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita M. and Noda M. (2001). Roles of septins in the mammalian cytokinesis machinery. Cell Struct. Funct. 26, 667-670. 10.1247/csf.26.667 [DOI] [PubMed] [Google Scholar]
- Kotloff K. L., Winickoff J. P., Ivanoff B., Clemens J. D., Swerdlow D. L., Sansonetti P. J., Adak G. K. and Levine M. M. (1999). Global burden of Shigella infections: implications for vaccine development and implementation of control strategies. Bull. WHO 77, 651-666. [PMC free article] [PubMed] [Google Scholar]
- Kotloff K. L., Riddle M. S., Platts-Mills J. A., Pavlinac P. and Zaidi A. K. M. (2017). Shigellosis. Lancet. doi:10.1016/0140-6736(17)33296-33298 [Epub ahead of print] 10.1016/0140-6736(17)33296-33298 [DOI] [PubMed] [Google Scholar]
- Krokowski S. and Mostowy S. (2016). Interactions between Shigella flexneri and the autophagy machinery. Front. Cell Infect. Microbiol. 6, 17 10.3389/fcimb.2016.00017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuri P., Schieber N. L., Thumberger T., Wittbrodt J., Schwab Y. and Leptin M. (2017). Dynamics of in vivo ASC speck formation. J. Cell Biol. 216, 2891-2909. 10.1083/jcb.201703103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam P., Harvie E. A. and Huttenlocher A. (2013). Heat shock modulates neutrophil motility in zebrafish. PLoS ONE 8, e84436 10.1371/journal.pone.0084436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landsverk M. L., Weiser D. C., Hannibal M. C. and Kimelman D. (2010). Alternative splicing of sept9a and sept9b in zebrafish produces multiple mRNA transcripts expressed throughout development. PLoS ONE 5, e10712 10.1371/journal.pone.0010712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurenti E. and Göttgens B. (2018). From haematopoietic stem cells to complex differentiation landscapes. Nature 553, 418-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P. P., Lobato-Márquez D., Pramanik N., Sirianni A., Daza-Cajigal V., Rivers E., Cavazza A., Bouma G., Moulding D., Hultenby K. et al. (2017). Wiskott-Aldrich Syndrome protein (WASp) regulates autophagy and inflammasome activity in innate immune cells. Nat. Commun. 8, 1576 10.1038/s41467-017-01676-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B., Mizushima N. and Virgin H. W. (2011). Autophagy in immunity and inflammation. Nature 469, 323-335. 10.1038/nature09782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levraud J.-P., Palha N., Langevin C. and Boudinot P. (2014). Through the looking glass: witnessing host-virus interplay in zebrafish. Trends Microbiol. 22, 490-497. 10.1016/j.tim.2014.04.014 [DOI] [PubMed] [Google Scholar]
- Li M., Zhao L., Page-McCaw P. S. and Chen W. (2016). Zebrafish genome engineering using the CRISPR-Cas9 system. Trends Genet. 32, 815-827. 10.1016/j.tig.2016.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Jiang W., Yu Q., Liu W., Zhou P., Li J., Xu J., Xu B., Wang F. and Shao F. (2017). Ubiquitination and degradation of GBPs by a Shigella effector to suppress host defence. Nature 551, 378-383. 10.1038/nature24467 [DOI] [PubMed] [Google Scholar]
- Lieschke G. J. and Currie P. D. (2007). Animal models of human disease: zebrafish swim into view. Nat. Rev. Genet. 8, 353-367. 10.1038/nrg2091 [DOI] [PubMed] [Google Scholar]
- Lieschke G. J. and Trede N. S. (2009). Fish immunology . Curr. Biol. 19, R678-R682. 10.1016/j.cub.2009.06.068 [DOI] [PubMed] [Google Scholar]
- Lima I. F. N., Havt A. and Lima A. A. M. (2015). Update on molecular epidemiology of Shigella infection. Curr. Opin. Gastroenterol. 31, 30-37. 10.1097/MOG.0000000000000136 [DOI] [PubMed] [Google Scholar]
- Long Y., Li L., Li Q., He X. and Cui Z. (2012). Transcriptomic characterization of temperature stress responses in larval zebrafish. PLoS ONE 7, e37209 10.1371/journal.pone.0037209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madigan C. A., Cameron J. and Ramakrishnan L. (2017). A zebrafish model of Mycobacterium leprae granulomatous infection. J. Infect. Dis. 216, 776-779. 10.1093/infdis/jix329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik A. and Kanneganti T.-D. (2017). Inflammasome activation and assembly at a glance. J. Cell Sci. 130, 3955-3963. 10.1242/jcs.207365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani S., Wierzba T. and Walker R. I. (2016). Status of vaccine research and development for Shigella. Vaccine 34, 2887-2894. 10.1016/j.vaccine.2016.02.075 [DOI] [PubMed] [Google Scholar]
- Manz M. G. and Boettcher S. (2014). Emergency granulopoiesis. Nat. Rev. Immunol. 14, 302-314. 10.1038/nri3660 [DOI] [PubMed] [Google Scholar]
- Masud S., Torraca V. and Meijer A. H. (2017). Modeling infectious diseases in the context of a developing immune system. Curr. Top. Dev. Biol. 124, 277-329. 10.1016/bs.ctdb.2016.10.006 [DOI] [PubMed] [Google Scholar]
- Mazon-Moya M. J., Willis A. R., Torraca V., Boucontet L., Shenoy A. R., Colucci-Guyon E. and Mostowy S. (2017). Septins restrict inflammation and protect zebrafish larvae from Shigella infection. PLoS Pathog. 13, e1006467 10.1371/journal.ppat.1006467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon M. B., Sawada A., Chaturvedi A., Mishra P., Schuster-Gossler K., Galla M., Schambach A., Gossler A., Förster R., Heuser M. et al. (2014). Genetic deletion of SEPT7 reveals a cell type-specific role of septins in microtubule destabilization for the completion of cytokinesis. PLoS Genet. 10, e1004558 10.1371/journal.pgen.1004558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao E. A., Rajan J. V. and Aderem A. (2011). Caspase-1 induced pyroptotic cell death. Immun. Rev. 243, 206-214. 10.1111/j.1600-065X.2011.01044.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostowy S. (2014). Multiple roles of the cytoskeleton in bacterial autophagy. PLoS Pathog. 10, e1004409 10.1371/journal.ppat.1004409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostowy S. and Cossart P. (2009). Cytoskeleton rearrangements during Listeria infection: Clathrin and septins as new players in the game. Cell Motil. Cytoskeleton 66, 816-823. 10.1002/cm.20353 [DOI] [PubMed] [Google Scholar]
- Mostowy S. and Cossart P. (2012). Septins: the fourth component of the cytoskeleton. Nat. Rev. Mol. Cell Biol. 13, 183-194. [DOI] [PubMed] [Google Scholar]
- Mostowy S. and Shenoy A. R. (2015). The cytoskeleton in cell-autonomous immunity: structural determinants of host defence. Nat. Rev. Immunol. 15, 559-573. 10.1038/nri3877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostowy S., Nam Tham T., Danckaert A., Guadagnini S., Boisson-Dupuis S., Pizarro-Cerdá J. and Cossart P. (2009a). Septins regulate bacterial entry into host cells. PLoS ONE 4, e4196 10.1371/journal.pone.0004196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostowy S., Danckaert A., Tham T. N., Machu C., Guadagnini S., Pizarro-Cerdá J. and Cossart P. (2009b). Septin 11 restricts InlB-mediated invasion by Listeria . J. Biol. Chem. 284, 11613-11621. 10.1074/jbc.M900231200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostowy S., Bonazzi M., Hamon M. A., Tham T. N., Mallet A., Lelek M., Gouin E., Demangel C., Brosch R., Zimmer C. et al. (2010). Entrapment of intracytosolic bacteria by septin cage-like structures. Cell Host Microbe 8, 433-444. [DOI] [PubMed] [Google Scholar]
- Mostowy S., Sancho-Shimizu V., Hamon M. A., Simeone R., Brosch R., Johansen T. and Cossart P. (2011). p62 and NDP52 proteins target intracytosolic Shigella and Listeria to different autophagy pathways. J. Biol. Chem. 286, 26987-26995. 10.1074/jbc.M111.223610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostowy S., Boucontet L., Mazon Moya M. J., Sirianni A., Boudinot P., Hollinshead M., Cossart P., Herbomel P., Levraud J.-P. and Colucci-Guyon E. (2013). The zebrafish as a new model for the in vivo study of Shigella flexneri interaction with phagocytes and bacterial autophagy. PLoS Pathog. 9, e1003588 10.1371/journal.ppat.1003588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigro G., Rossi R., Commere P.-H., Jay P. and Sansonetti P. J. (2014). The cytosolic bacterial peptidoglycan sensor Nod2 affords stem cell protection and links microbes to gut epithelial regeneration. Cell Host Microbe 15, 792-798. 10.1016/j.chom.2014.05.003 [DOI] [PubMed] [Google Scholar]
- Nigro G., Hanson M., Fevre C., Lecuit M. and Sansonetti P. J. (2016). Intestinal organoids as a novel tool to study microbes-epithelium interactions. In Methods in Molecular Biology, pp. 1-12. Clifton, NJ: Humana Press; 10.1007/7651_2016_12 [DOI] [PubMed] [Google Scholar]
- Oehlers S. H., Cronan M. R., Scott N. R., Thomas M. I., Okuda K. S., Walton E. M., Beerman R. W., Crosier P. S. and Tobin D. M. (2015). Interception of host angiogenic signalling limits mycobacterial growth. Nature 517, 612-615. 10.1038/nature13967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M., Yoshimori T., Suzuki T., Sagara H., Mizushima N. and Sasakawa C. (2005). Escape of intracellular Shigella from autophagy. Science 307, 727-731. 10.1126/science.1106036 [DOI] [PubMed] [Google Scholar]
- Pelegrin P. and Surprenant A. (2009). Dynamics of macrophage polarization reveal new mechanism to inhibit IL-1β release through pyrophosphates. EMBO J. 28, 2114-2127. 10.1038/emboj.2009.163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendleton J. N., Gorman S. P. and Gilmore B. F. (2013). Clinical relevance of the ESKAPE pathogens. Expert Rev. Anti. Infect. Ther. 11, 297-308. 10.1586/eri.13.12 [DOI] [PubMed] [Google Scholar]
- Perdomo O. J., Cavaillon J. M., Huerre M., Ohayon H., Gounon P. and Sansonetti P. J. (1994). Acute inflammation causes epithelial invasion and mucosal destruction in experimental shigellosis. J. Exp. Med. 180, 1307-1319. 10.1084/jem.180.4.1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phalipon A. and Sansonetti P. J. (2007). Shigella’s ways of manipulating the host intestinal innate and adaptive immune system: a tool box for survival? Immunol. Cell Biol. 85, 119-129. 10.1038/sj.icb7100025 [DOI] [PubMed] [Google Scholar]
- Phalipon A., Mulard L. A. and Sansonetti P. J. (2008). Vaccination against shigellosis: is it the path that is difficult or is it the difficult that is the path? Microbes Infect. 10, 1057-1062. 10.1016/j.micinf.2008.07.016 [DOI] [PubMed] [Google Scholar]
- Picking W. D. and Picking W. L. (2016). Shigella: Molecular and Cellular Biology. Lawrence, KS: Caister Academic Press. [Google Scholar]
- Renshaw S. A. and Trede N. S. (2012). A model 450 million years in the making: zebrafish and vertebrate immunity. Dis. Model. Mech. 5, 38-47. 10.1242/dmm.007138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saarikangas J. and Barral Y. (2011). The emerging functions of septins in metazoans. EMBO Rep. 12, 1118-1126. 10.1038/embor.2011.193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salgado-Pabón W., Konradt C., Sansonetti P. J. and Phalipon A. (2014). New insights into the crosstalk between Shigella and T lymphocytes. Trends Microbiol. 22, 192-198. 10.1016/j.tim.2014.02.002 [DOI] [PubMed] [Google Scholar]
- Schnupf P. and Sansonetti P. J. (2012). Quantitative RT-PCR profiling of the rabbit immune response: assessment of acute Shigella flexneri infection. PLoS ONE 7, e36446 10.1371/journal.pone.0036446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sereny B. (1955). Experimental Shigella keratoconjunctivitis; a preliminary report. Acta Microbiol. Acad. Sci. Hung. 2, 293-296. [PubMed] [Google Scholar]
- Shatzkes K., Singleton E., Tang C., Zuena M., Shukla S., Gupta S., Dharani S., Onyile O., Rinaggio J., Connell N. D. et al. (2016). Predatory bacteria attenuate Klebsiella pneumoniae burden in rat lungs. mBio. 7, e01847-16 10.1128/mBio.01847-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shatzkes K., Tang C., Singleton E., Shukla S., Zuena M., Gupta S., Dharani S., Rinaggio J., Connell N. D. and Kadouri D. E. (2017). Effect of predatory bacteria on the gut bacterial microbiota in rats. Sci. Rep. 7, 43483 10.1038/srep43483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim D.-H., Suzuki T., Chang S.-Y., Park S.-M., Sansonetti P. J., Sasakawa C. and Kweon M.-N. (2007). New animal model of shigellosis in the Guinea pig: its usefulness for protective efficacy studies. J. Immunol. 178, 2476-2482. 10.4049/jimmunol.178.4.2476 [DOI] [PubMed] [Google Scholar]
- Sirianni A., Krokowski S., Lobato-Márquez D., Buranyi S., Pfanzelter J., Galea D., Willis A., Culley S., Henriques R., Larrouy-Maumus G. et al. (2016). Mitochondria mediate septin cage assembly to promote autophagy of Shigella. EMBO Rep. 17, 1029-1043. 10.15252/embr.201541832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorbara M. T., Ellison L. K., Ramjeet M., Travassos L. H., Jones N. L., Girardin S. E. and Philpott D. J. (2013). The protein ATG16L1 suppresses inflammatory cytokines induced by the intracellular sensors Nod1 and Nod2 in an autophagy-independent manner. Immunity 39, 858-873. 10.1016/j.immuni.2013.10.013 [DOI] [PubMed] [Google Scholar]
- Stachura D. L., Svoboda O., Campbell C. A., Espín-Palazón R., Lau R. P., Zon L. I., Bartůněk P. and Traver D. (2013). The zebrafish granulocyte colony-stimulating factors (Gcsfs): 2 paralogous cytokines and their roles in hematopoietic development and maintenance. Blood 122, 3918-3928. 10.1182/blood-2012-12-475392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Standing A. S. I., Malinova D., Hong Y., Record J., Moulding D., Blundell M. P., Nowak K., Jones H., Omoyinmi E., Gilmour K. C. et al. (2017). Autoinflammatory periodic fever, immunodeficiency, and thrombocytopenia (PFIT) caused by mutation in actin-regulatory gene WDR1. J. Exp. Med. 214, 59-71. 10.1084/jem.20161228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockhammer O. W., Zakrzewska A., Hegedûs Z., Spaink H. P. and Meijer A. H. (2009). Transcriptome profiling and functional analyses of the zebrafish embryonic innate immune response to Salmonella infection. J. Immunol. 182, 5641-5653. 10.4049/jimmunol.0900082 [DOI] [PubMed] [Google Scholar]
- Stones D. H., Fehr A. G. J., Thompson L., Rocha J., Perez-Soto N., Madhavan V. T. P., Voelz K. and Krachler A. M. (2017). Zebrafish (Danio rerio) as a vertebrate model host to study colonisation, pathogenesis, and transmission of foodborne Escherichia coli O157. mSphere 2, e00365-17 10.1128/mSphereDirect.00365-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storisteanu D. M. L., Pocock J. M., Cowburn A. S., Juss J. K., Nadesalingam A., Nizet V. and Chilvers E. R. (2017). Evasion of neutrophil extracellular traps by respiratory pathogens. Am. J. Respir. Cell Mol. Biol. 56, 423-431. 10.1165/rcmb.2016-0193PS [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan C., Matty M. A., Jurczyszak D., Gabor K. A., Millard P. J., Tobin D. M. and Kim C. H. (2017). Infectious disease models in zebrafish. Methods Cell Biol. 138, 101-136. 10.1016/bs.mcb.2016.10.005 [DOI] [PubMed] [Google Scholar]
- Suzuki S., Suzuki T., Mimuro H., Mizushima T. and Sasakawa C. (2017). Shigella hijacks the glomulin-cIAPs-inflammasome axis to promote inflammation. EMBO Rep. 19, 89-101. 10.15252/embr.201643841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tattoli I., Sorbara M. T., Vuckovic D., Ling A., Soares F., Carneiro L. A. M., Yang C., Emili A., Philpott D. J. and Girardin S. E. (2012). Amino acid starvation induced by invasive bacterial pathogens triggers an innate host defense program. Cell Host Microbe 11, 563-575. 10.1016/j.chom.2012.04.012 [DOI] [PubMed] [Google Scholar]
- Teng T.-S., Ji A.-L., Ji X.-Y. and Li Y.-Z. (2017). Neutrophils and immunity: from bactericidal action to being conquered. J. Immun. Res. 2017, 9671604 10.1155/2017/9671604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurston T. L. M., Wandel M. P., von Muhlinen N., Foeglein Á. and Randow F. (2012). Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature 482, 414-418. 10.1038/nature10744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobin D. M., May R. C. and Wheeler R. T. (2012). Zebrafish: a see-through host and a fluorescent toolbox to probe host–pathogen interaction. PLoS Pathog. 8, e1002349 10.1371/journal.ppat.1002349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torraca V. and Mostowy S. (2016). Septins and bacterial infection. Front. Cell Dev. Biol. 4, 127 10.3389/fcell.2016.00127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torraca V. and Mostowy S. (2017). Zebrafish infection: from pathogenesis to cell biology. Trends. Cell. Biol. 28, 143-156. 10.1016/j.tcb.2017.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyrkalska S. D., Candel S., Angosto D., Gómez-Abellán V., Martín-Sánchez F., García-Moreno D., Zapata-Pérez R., Sánchez-Ferrer A., Sepulcre M. P., Pelegrín P. et al. (2016). Neutrophils mediate Salmonella Typhimurium clearance through the GBP4 inflammasome-dependent production of prostaglandins. Nat. Commun. 7, 12077 10.1038/ncomms12077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson J. and Sockett R. E. (2017). Predatory bacteria: moving from curiosity towards curative. Rev. Trends Microbiol. 25, 90-91. 10.1016/j.tim.2016.12.011 [DOI] [PubMed] [Google Scholar]
- van der Vaart M., Korbee C. J., Lamers G. E. M., Tengeler A. C., Hosseini R., Haks M. C., Ottenhoff T. H. M., Spaink H. P. and Meijer A. H. (2014). The DNA damage-regulated autophagy modulator DRAM1 links mycobacterial recognition via TLR-MYD88 to autophagic defense. Cell Host Microbe 15, 753-767. 10.1016/j.chom.2014.05.005 [DOI] [PubMed] [Google Scholar]
- Varela M., Figueras A. and Novoa B. (2017). Modelling viral infections using zebrafish: innate immune response and antiviral research. Antiviral Res. 139, 59-68. 10.1016/j.antiviral.2016.12.013 [DOI] [PubMed] [Google Scholar]
- Vincent W. J. B., Freisinger C. M., Lam P.-Y., Huttenlocher A. and Sauer J.-D. (2016). Macrophages mediate flagellin induced inflammasome activation and host defense in zebrafish. Cell Microbiol. 18, 591-604. 10.1111/cmi.12536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandel M. P., Pathe C., Werner E. I., Ellison C. J., Boyle K. B., von der Malsburg A., Rohde J. and Randow F. (2017). Gbps inhibit motility of Shigella flexneri but are targeted for degradation by the bacterial ubiquitin ligase IpaH9.8. Cell Host Microbe 22, 507-518.e5. 10.1016/j.chom.2017.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner A., Mellouk N., Lopez-Montero N., Chang Y.-Y., Souque C., Schmitt C. and Enninga J. (2016). Macropinosomes are key players in early Shigella invasion and vacuolar escape in epithelial cells. PLoS Pathog. 12, e1005602 10.1371/journal.ppat.1005602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch M. D. and Way M. (2013). Arp2/3-mediated actin-based motility: a tail of pathogen abuse. Cell Host Microbe 14, 242-255. 10.1016/j.chom.2013.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willingham S. B., Bergstralh D. T., O'Connor W., Morrison A. C., Taxman D. J., Duncan J. A., Barnoy S., Venkatesan M. M., Flavell R. A., Deshmukh M. et al. (2007). Microbial pathogen-induced necrotic cell death mediated by the inflammasome components CIAS1/Cryopyrin/NLRP3 and ASC. Cell Host Microbe 2, 147-159. 10.1016/j.chom.2007.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis A. R., Moore C., Mazon-Moya M., Krokowski S., Lambert C., Till R., Mostowy S. and Sockett R. E. (2016). Injections of predatory bacteria work alongside host immune cells to treat Shigella infection in zebrafish larvae. Curr. Biol. 26, 3343-3351. 10.1016/j.cub.2016.09.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (2017). Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. http://www.who.int/medicines/publications/WHO-PPL-Short_Summary_25Feb-ET_NM_WHO.pdf.
- Yadav S., Oses-Prieto J. A., Peters C. J., Zhou J., Pleasure S. J., Burlingame A. L., Jan L. Y. and Jan Y.-N. (2017). TAOK2 kinase mediates PSD95 stability and dendritic spine maturation through Septin7 phosphorylation. Neuron 93, 379-393. 10.1016/j.neuron.2016.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. and Klionsky D. J. (2010). Eaten alive: a history of macroautophagy. Nat. Cell Biol. 12, 814-822. 10.1038/ncb0910-814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J.-Y., Lee S.-N., Chang S.-Y., Ko H.-J., Ryu S. and Kweon M.-N. (2014). A mouse model of shigellosis by intraperitoneal infection. J. Infect. Dis. 209, 203-215. 10.1093/infdis/jit399 [DOI] [PubMed] [Google Scholar]
- Yoshida N., Frickel E.-M. and Mostowy S. (2017). Macrophage–microbe interactions: lessons from the zebrafish model. Front. Immunol. 8, 1703 10.3389/fimmu.2017.01703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai G., Gu Q., He J., Lou Q., Chen X., Jin X., Bi E. and Yin Z. (2014). Sept6 is required for ciliogenesis in Kupffer's vesicle, the pronephros, and the neural tube during early embryonic development. Mol. Cell. Biol. 34, 1310-1321. 10.1128/MCB.01409-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zychlinsky A., Prevost M. C. and Sansonetti P. J. (1992). Shigella flexneri induces apoptosis in infected macrophages. Nature 358, 167-169. 10.1038/358167a0 [DOI] [PubMed] [Google Scholar]