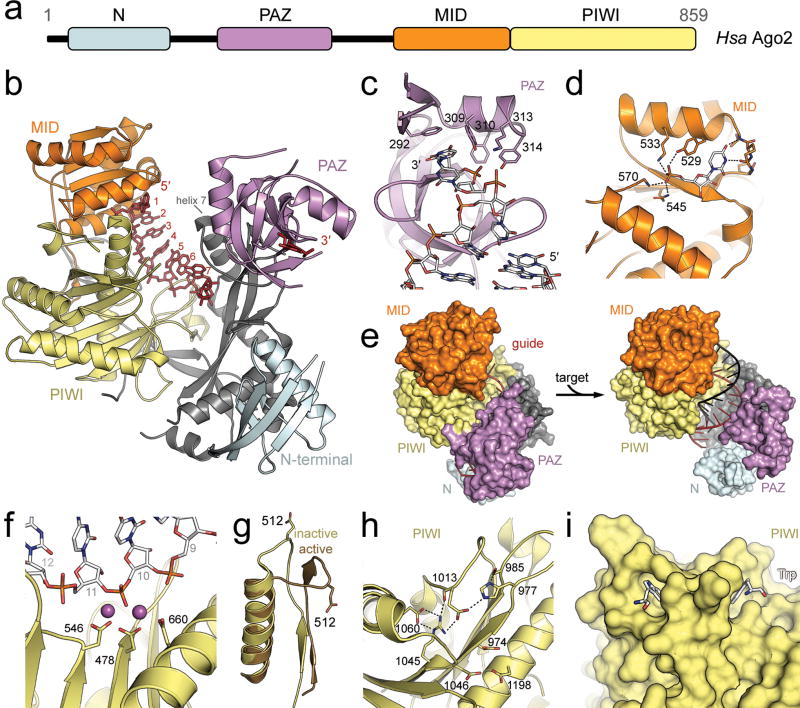
Figure 5.
Snapshots of RISC in action. (a) The domain structure of Argonaute is relatively well conserved in the AGO clade. PIWI clade proteins lack the N and PAZ domains. (b) The crystal structure of human Ago2 bound to an RNA guide strand (PDB ID: 4EI1). The seed region (nt 2–6) is pre-arranged in A-form geometry while the downstream portions of strand cannot be modeled due to disorder, as is typical in the absence of a target strand. Helix 7 is observed to rest against guide strand bases 6 and 7, distorting their geometry and providing an apparent barrier to target binding that is presumably circumvented via a conformational change. (c) The PAZ domain of human Ago2 recognizes the 3′-terminal 2 nt overhang typical of helices involved in RNAi (PDB ID: 1SI3). Conserved residues contacting the 3′-terminus are represented as sticks and labeled. A hydrophobic pocket receives the terminal nucleobase. Nucleotides from the 5′-terminus can be seen in the bottom right, making only slight contact with the PAZ domain. (d) The MID domain is responsible for recognition of a phosphorylated 5′-terminus (PDB ID: 3LUJ). This UMP-bound crystal structure reveals the polar contacts that drive phosphate recognition as well as elucidating the base-specific contacts that grant the MID domain its preference for a 5′-terminal A or U. (e) A pair of T. thermophilus crystal structures illustrate a conformational change that results upon extensive base pairing between guide and target (PDB ID: left, 3DLH; right, 3HM9). RISC binding to a 19 nt target strand allows formation of an A-form helix that induces release of the guide strand’s 3′-terminus from the PAZ domain along with a drastic opening of the two Argonaute lobes. The target-bound model shows that the N domain blocks formation of a longer helix. (f) In the catalytic center of T. thermophilus Argonaute’s PIWI domain, three aspartic acid residues coordinate a pair of Mg2+ ions for cleavage of the target strand, shown as white sticks with nucleotides numbered in gray (PDB ID: 3HVR). (g) In T. thermophilus the target-induced conformation change also involves reorientation of L2, which contains a glutamic acid residue. If target binding is incomplete or absent, the inactive state is sampled (yellow). In the target-bound state (brown), the glutamic acid is deposited adjacent to the aforementioned catalytic triad, completing a tetrad typical of RNase H enzymes (PDB ID: bound to a 12 nt target and inactive, 3HO1; bound to a 19 nt target and active, 3HM9). (h) The pre-ordered catalytic tetrad as observed in the absence of target strand in K. polysporus Argonaute (PDB ID: 4F1N). Residue 1013 here corresponds to T. thermophilus residue 512. (i) The PIWI domain of human Ago2 bears two tryptophan binding sites that complement the side chain geometry expected from a GW protein binding partner (PDB ID: 4EI1). Free tryptophan was present in the crystallization conditions and the pair of bound amino acids is represented as sticks.