Figure 2.
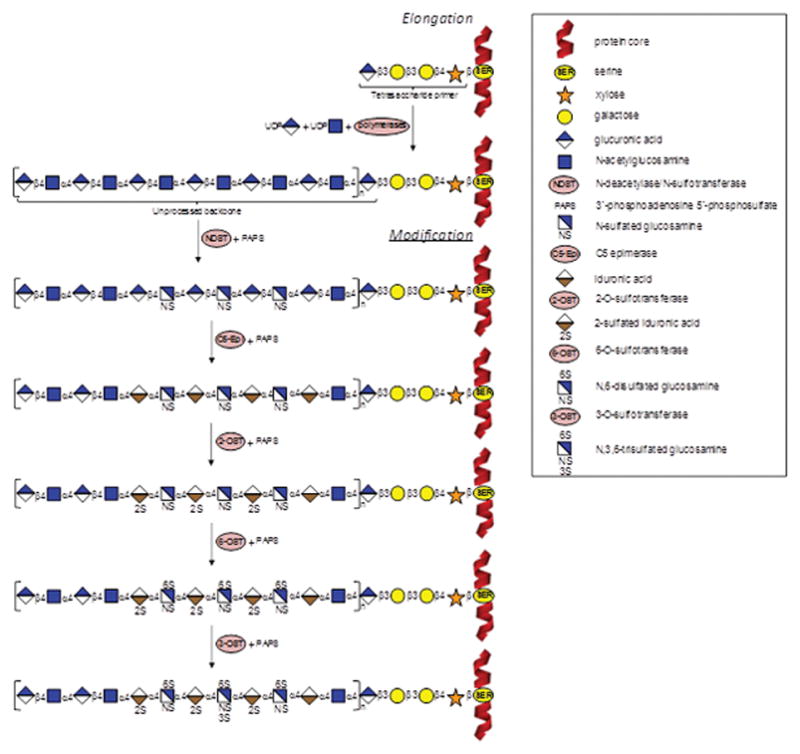
Schematic representation of heparin/heparan sulfate biosynthetic pathway. In the elongation stage, polymerization of the heparin/heparan sulfate chains is made by activities of polymerases (α-UDP-GlcNAc-glycosyltransferase and α-UDP-GlcA-glycosyltransferase) and thereafter by N-deacetylase/N-sulfotransferase (NDST) combined with the sulfate donor PAPS (3′-phosphoadenosine 5′-phosphosulfate). This process starts on a tetrasacchaide primer attached to a serine residue in the protein core. In the modification stage, C5-Epi (epimerase), 2-OST, 6-OST, and 3-OST (2-, 6- and 3-O-sulfotransferases) catalyze respectively the conversion of GlcA into IdoA, 2-sulfation at the IdoA, 6-sulfation at the GlcNS, and 3-sulfation at the GlcNS6S units. Heparin/heparan sulfate bears the same machinery of biosynthesis, varying only at the extensions of the reactions: heparin is much more processed than heparan sulfate.
