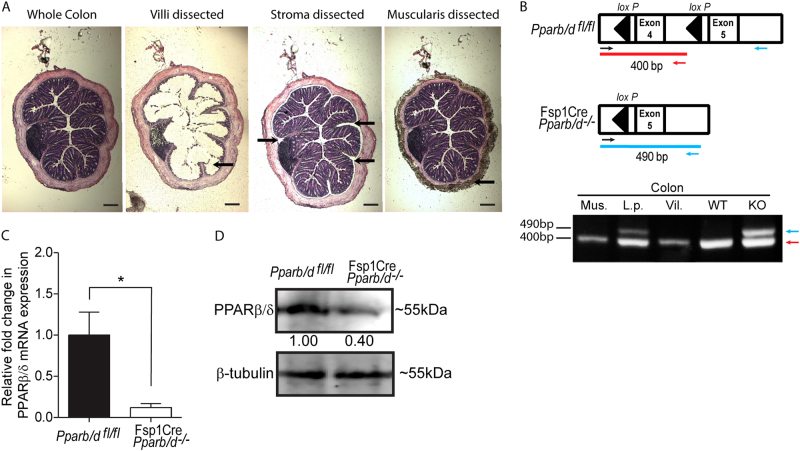
Fig. 1.
PPARβ/δ expression is ablated in colonic fibroblasts of FSPCre-Pparb/d−/− mice. a Representative H&E-stained images of colons before and after laser capture microdissection of colonic layers, mucosal, lamina propria, and muscularis. FSPCre-Pparb/d−/− mice were generated in-house by crossing Pparb/dfl/fl and FspCre mice progenies, and backcrossed with Pparb/dfl/fl for at least six generations. Scale: 500 µm (b) Schematic diagram depicting the relative position of PCR genotyping primers for floxed and deleted Pparb/d exon 4 alleles, at 400 bp (red), and 490 bp (blue), respectively (top). Sample DNA were processed as instructed in the KAPA Express Extract kit and 2 × KAPA2G Fast Genotyping Mix. DNA gel image showing genotyping results indicated tissue layers colonic submucosa and lamina propria layer (L.p.); mucosal villi (Vil.); smooth muscle (Mus.) layers. c Representative images showing the outgrowth of fibroblasts from colonic explants from Pparb/dfl/fl and FSPCre-Pparb/d−/− mice. Fresh colons were dissected longitudinally and washed in ice cold phosphate buffered solution (PBS) containing 30% antimycotic/antibiotic solution. Specimens were minced to 1–2 mm2 pieces and incubated in fresh culture media for 72 h to allow colonic fibroblasts to migrate out of the explants. Migrated cells were washed and cultured for another 48 h before protein and RNA analyses. Scale bar: 500 µm. c–d Relative fold change in expression of PPARβ/δ mRNA c and PPARβ/δ protein d in migrated colonic fibroblasts. Total RNA was extracted from cells using TRIzol Reagent with E.Z.N.A Total RNA Kit according to manufacturer’s protocol. Reverse transcription real-time quantitative PCR (RT-qPCR) were performed with 18S rRNA as a housekeeping gene. Samples were homogenized and lysed in mammalian protein extraction reagent (M-PER) supplemented with complete protease inhibitor mix. Far-infrared immunoblotting was performed. β-tubulin that served as housekeeping protein was from the same samples. Data are represented as mean ± S.E.M. from at least four independent experiments