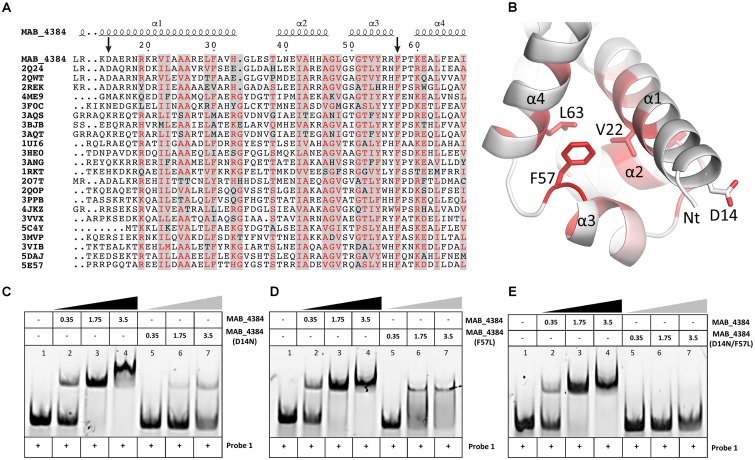
FIGURE 4.
D14N and F57L mutations abrogate binding of MAB_4384 to its palindromic DNA target. (A) Multiple primary sequence alignments of MAB_4384 N-terminus with other TetR family members whose crystal structures are known (PDB code indicated) from different microorganisms showing the conservation of the Asp14 and Phe57 residues (indicated with black arrows). (B) Mapping of the mutations on the crystal structure of MAB_4384, the degree of residue conservation (obtained from the sequence alignment in A) is represented by the coloration, from white (low conservation) to red (high conservation). (A,B) Were prepared using the ENDscript server (http://endscript.ibcp.fr/ESPript/ENDscript/). EMSA were performed using increasing concentrations (in μM) of the purified MAB_4384 (D14N) (C), MAB_4384 (F57L) (D) or MAB_4384 (D14N/F57L) (E) in the presence of Probe 1. Wild-type MAB_4384 was included as a positive control in each assay. The concentration of Probe 1 was fixed at 280 nM. Gel shifts were revealed by fluorescence emission. Three independent experiments were performed with similar results.