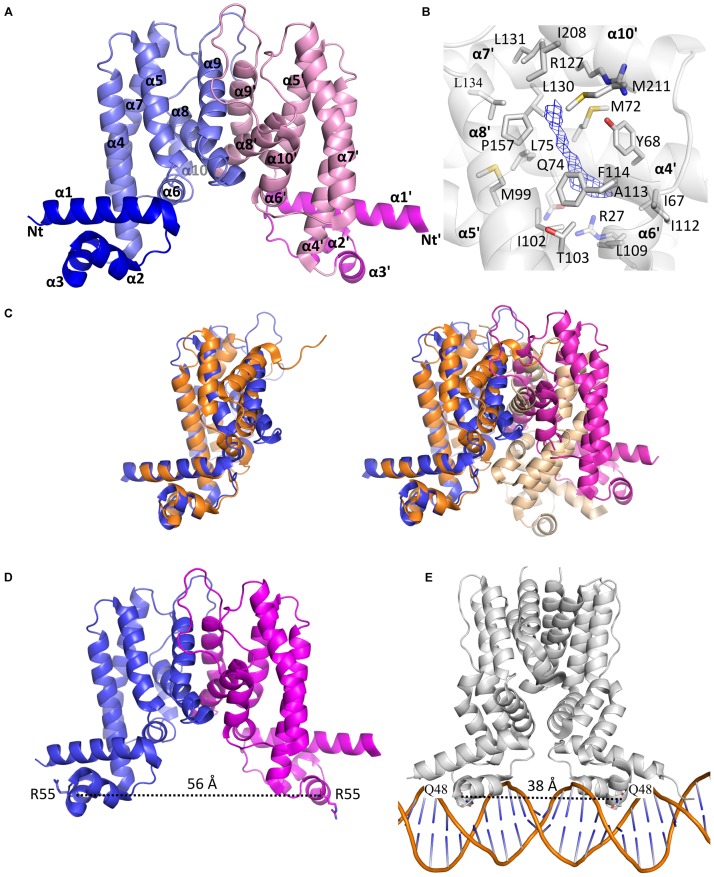
FIGURE 6.
The homodimeric crystal structure of MAB_4384. (A) Overall structure of the MAB_4384 dimer displayed as cartoon representation. The LBDs of each subunit are colored in slate and pink while the DNA binding domains are colored in blue and magenta. Helices are indicated by the α signs followed by numbers, Nt and Ct stands for N-terminus and C-terminus and ’ is for chain B. (B) Putative ligand binding pocket in the LBD of MAB_4384. The Fo-Fc simulated annealed omit map contoured at 3 σ level is shown in blue. Residues that are 4 Å around the electron density blob and that are potential amino acids of the ligand binding site are shown as sticks. (C) Structural comparison of MAB_4384 with the crystal structure of the M. smegmatis LfrR repressor (PDB id: 2V57). The left panel represents the superposition of one monomer of MAB_4384 (in blue) on one monomer of LfrR (in orange). The superposition of the two homodimers is shown on the right panel, the two subunits of MAB_4384 are in blue and magenta and the two monomers of LfrR are in orange and wheat. (D,E) The figures compare the distance between the two DNA binding domains in MAB_4384 (D) and in the crystal structure of the M. smegmatis TetR Ms6564 protein bound to its DNA target (PDB id: 4JL3).