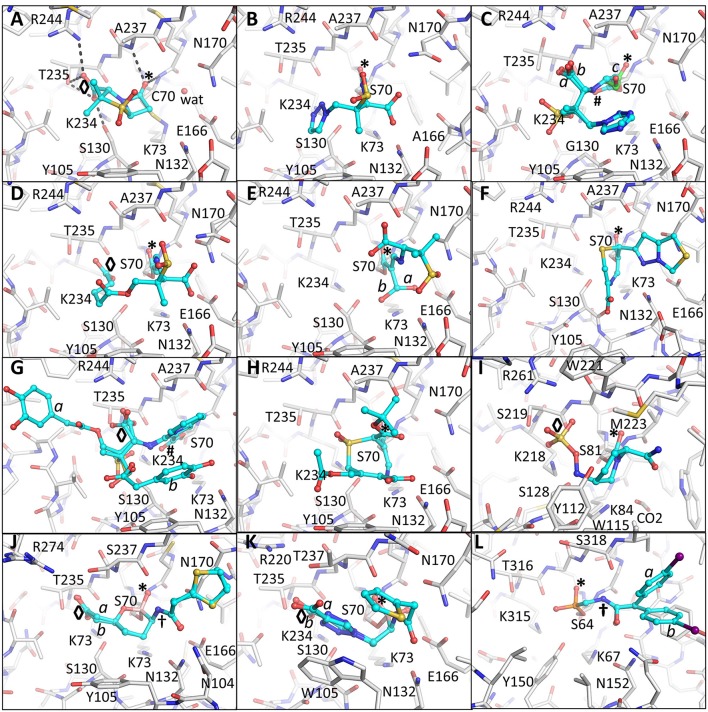
Figure 2.
Crystallographically determined binding modes of β-lactamase inhibitors. (A) Sulbactam bound in a pre-acylation/Michaelis-Menten binding mode in the S70C mutant of SHV-1 β-lactamase. The S70C mutations changes the reactivity of the catalytic S70 nucleophile; the C70 residue forms a covalent sulfonamide bond with the conserved K73 allowing capture of the pre-acylation complex. Hydrogen bonds between the carboxyl and carbonyl oxygens are depicted as dashed black lines. The occupied carboxyl pocket and oxyanion hole are labeled “◇” and “*”, respectively. These labels are used through subsequent panels of this figure where applicable. The deacylation water is shown as a solid red sphere labeled “wat;” (B) tazobactam, in the trans-enamine conformation, bound to the deacylation deficient E166A mutant of SHV-1; (C) tazobactam, in the cis-enamine conformation, bound to the inhibitor-resistant S130G mutant of SHV-1. Tazobactam adopts three conformations two of which are cis-enamine (0.33 occupancy with cyan carbon atoms each labeled “a” and “b”) and a fragmented species with green carbon atoms labeled “c” (also 0.33 occupancy). These labels for alternate conformations are used when needed in subsequent panels of this Figure. The cis-enamine and fragmented species have their carbonyl oxygens positioned outside (labeled “#”) and inside the oxyanion hole (labeled “*”), respectively; (D) SA2-13 complexed to SHV-1; (E) PSR-2-283A complexed to SHV-1. The hydroxymethyl moiety was observed to be in two conformations (labeled “a” and “b”). The major conformation hydrogen bonds with the deacylation water (not shown) and the second conformation does not. Residue S130 is also in two conformations; (F) penem 1 bound to SHV-1; (G) LN-1-255 complexed to SHV-1. Two conformations of the tail of LN-1-255 are observed (“a” and “b”); (H) DCM-1-10 bound to SHV-1; (I) Avibactam bound to Class D OXA-24 β-lactamase; (J) Vaborbactam complexed to CTX-M-15. Two conformations for vaborbactam were observed. The amide moiety of vaborbactam (labeled “†”) makes hydrogen bonds across the active site groove; (K) S02030 bound to KPC-2. Two conformations were observed for the carboxyl-triazole moiety (labeled “a” and “b”); (L) Phosphonate 3 complexed to P99 β-lactamase. The iodobenzene ring was present in two conformations. Like in vaborbactam, the amide moiety of phosphonate 3 makes hydrogen bonds across the active site.