Figure 5.
ZUFSP Catalytic Activity Requires the UBZ Domain
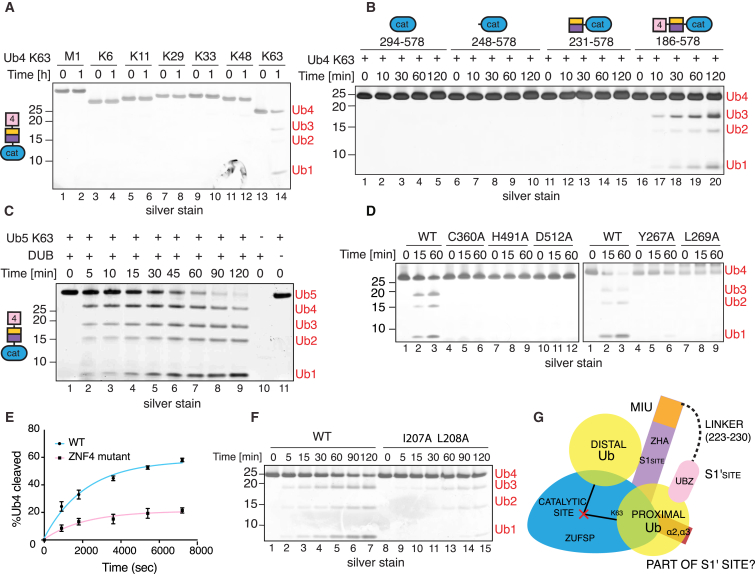
(A) DUB assay of ZUFSPZNF4-MIU-cat. Purified protein was incubated with each Ub4 chain type for the indicated time points, and reaction products were separated by SDS-PAGE and silver stained.
(B) DUB assay as in (A) comparing DUB activity of the indicated ZUFSP constructs.
(C) DUB assay monitoring K63-pentaUb cleavage by ZUFSPZNF4-MIU-Cat, with reaction products visualized as in (A).
(D) DUB assay comparing activity of the indicated ZUFSP mutants at cleaving K63-tetraUb
(E) Comparison of DUB activities of ZUFSPZNF4-MIU-Cat with UBZ mutant that cannot bind ubiquitin. Percentage of cleaved tetraUb was quantified from Sypro Ruby-stained gels. Data from three independent experiments were fitted using nonlinear regression, one phase exponential decay. SD error bars are shown.
(F) DUB assay as in (D) monitoring cleavage of K63-linked tetraUb.
(G) Model depicting substrate binding and catalysis in ZUFSP. The distal Ub is stabilized by contacts with the ZHA domain (violet) and the catalytic core (blue). The polyUb binding UBZ domain (ZNF4) (pink) is required for catalytic activity and we suggest that it may form the proximal Ub binding S1’ site.
See also Figure S5.