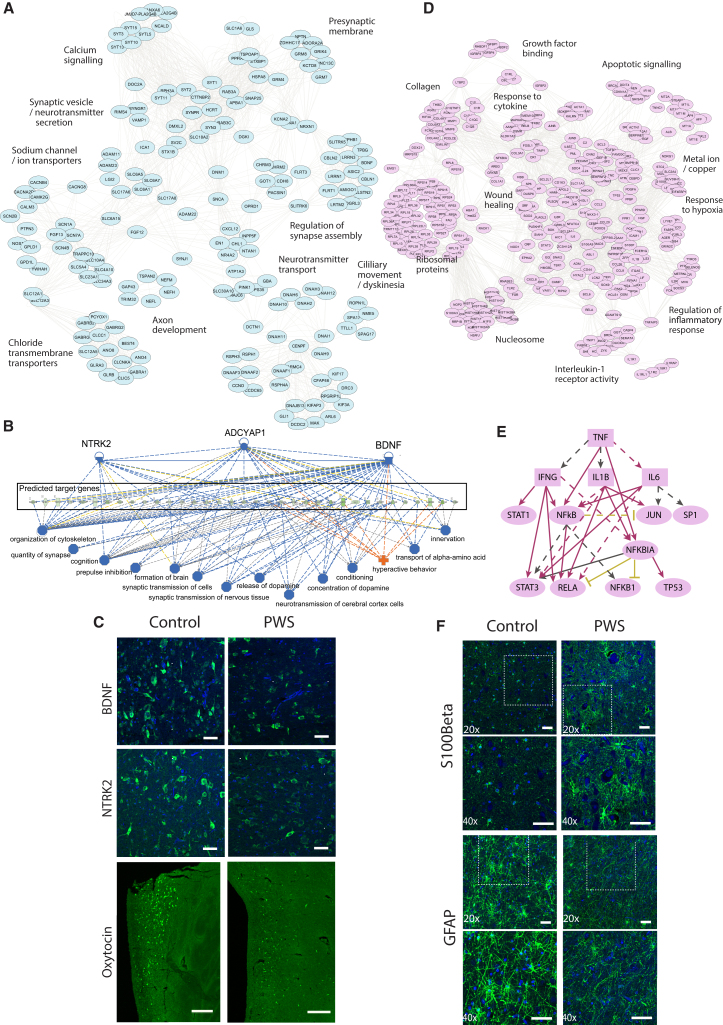
Figure 3.
Pathways Predicted to Be Affected by Changes in Gene Expression Seen in PWS Hypothalamus
(A) A gene annotation network illustrating terms (Gene Ontology, Reactome, Key) enriched among downregulated DEGs. Nodes represent downregulated DEGs annotated with illustrated terms; edges join pairs of genes annotated with the respective term.
(B) Ingenuity Pathway Analysis (IPA) regulator effects analysis indicates the inhibition of regulatory factors NTRK2, ADCYAP1, and BDNF (top) with predicted effects on target genes and processes. Phenotypes predicted to occur as a consequence of the gene expression changes are shown in blue (inhibited) or orange (enhanced).
(C) Representative FISH images of BDNF and NTRK2 mRNA-expressing cells in the ventromedial nucleus of the hypothalamus and oxytocin mRNA-expressing cells in the paraventricular nucleus of the hypothalamus in PWS and control samples (BDNF [n = 2 PWS, n = 2 controls], NTRK2 [n = 2 PWS, n = 1 control], and oxytocin [n = 2 PWS, n = 1 control]).
(D) A gene annotation network illustrating terms enriched among upregulated DEGs. Nodes and edges as in Figure 2A.
(E) IPA upstream regulator analysis indicates inhibition of TNF/NFKb signaling.
(F) Representative immunohistochemistry images of S100Beta- and GFAP-immunoreactive cells in the ventromedial nucleus of the hypothalamus in PWS and control samples.