Figure 1.
WIPI2 Interacts with RAB11A
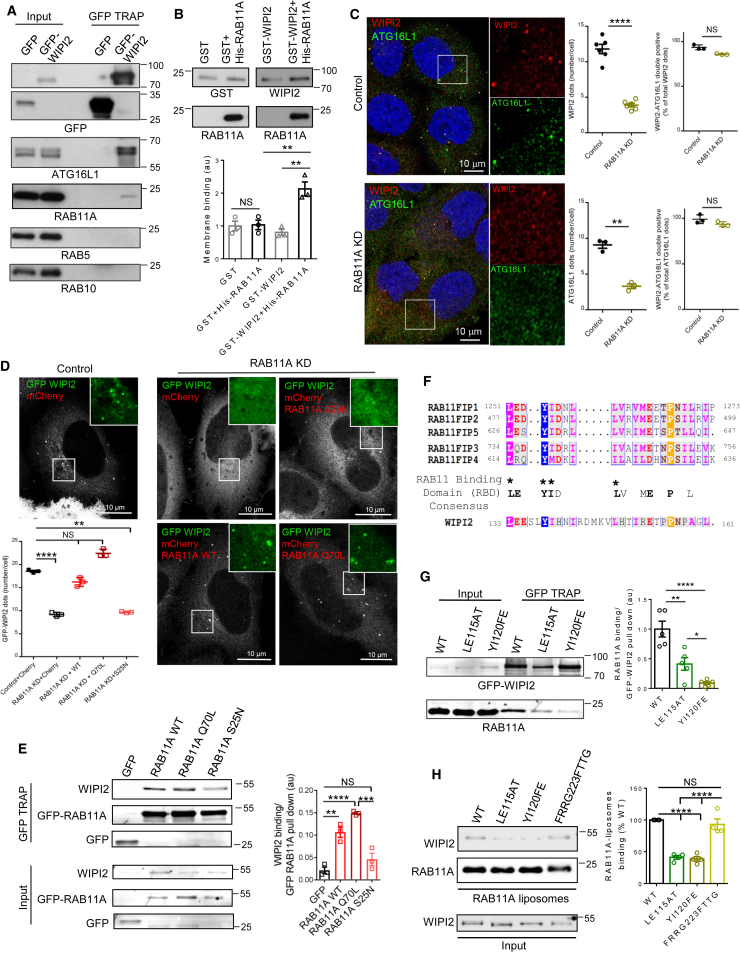
(A) GFP-WIPI2 and GFP were immuno-precipitated using GFP-TRAP on lysates from HeLa cells stably expressing GFP-WIPI2 or GFP-empty vector under starvation conditions (EBSS) for 2 hr; blots were probed as indicated.
(B) Binding of GST-WIPI2 to RAB11A-liposomes was analyzed by probing for liposome-bound GST (GST antibody lanes 1–2) or GST-WIPI2 (WIPI2 antibody, lanes 3–4) (see STAR Methods). Data are means ± SEM, n = 3; One-way ANOVA with post hoc Tukey's test, ∗∗p < 0.01; NS, not significant.
(C) HeLa cells treated with control or RAB11A siRNA, starved for 2 hr, and labeled for WIPI2 and ATG16L1. Quantification of WIPI2-, ATG16L1-single-positive structures (number/cell) is shown. WIPI2-ATG16L1 double-positive structures are expressed as percentage of total WIPI2 or ATG16L1 structures. Data are means ± SEM (n = 6 for WIPI2, n = 3 for ATG16L1, 50 cells per condition); two-tailed paired t test, ∗∗p < 0.01, ∗∗∗∗p < 0.0001; NS, not significant.
(D) HeLa cells treated as in (A) transfected with GFP-WIPI2 in combination with mCherry-empty or mCherry-RAB11A WT or mutants. Quantification of WIPI2 structures/cell is shown. Data are means ± SEM (n = 3, 40 cells per condition); One-way ANOVA with post hoc Tukey's test, ∗∗p < 0.01, ∗∗∗∗p < 0.0001; NS, not significant.
(E) HeLa cells transfected with GFP-RAB11A WT or mutants were starved for 1 hr and processed for GFP-TRAP. The amount of WIPI2 pull-down by RAB11A WT and mutants is shown. Data are means ± SEM (n = 3 independent experiments); one-way ANOVA with post hoc Tukey's test, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001; NS, not significant.
(F) Alignment of amino acid sequence of the RAB11-binding domain (RBD) of RAB11FIPs with WIPI2 (isoform B, residues 133–161). Colored box, white character strict identity. Consensus sequence for RAB11 binding is shown; hydrophobic residues forming a RAB11-binding patch are marked with asterisks.
(G) HeLa cells transfected with GFP-WIPI2 WT, GFP-WIPI2 LE115AT and GFP-WIPI2 YI120FE and starved for 2 hr were processed for GFP-TRAP as in (A). Data are means ± SEM, n = 5; one-way ANOVA, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.0001.
(H) Binding of WT, LE115AT, YI120FE, and FRRG223FTTG WIPI2-FLAG recombinant proteins to RAB11A-containing liposomes was measured in vitro by liposome sedimentation assay (see STAR Methods). WIPI2 association with liposomes is shown. Data are means ± SEM, n = 4; one-way ANOVA with post hoc Tukey's test, ∗∗∗∗p < 0.0001; NS, not significant.