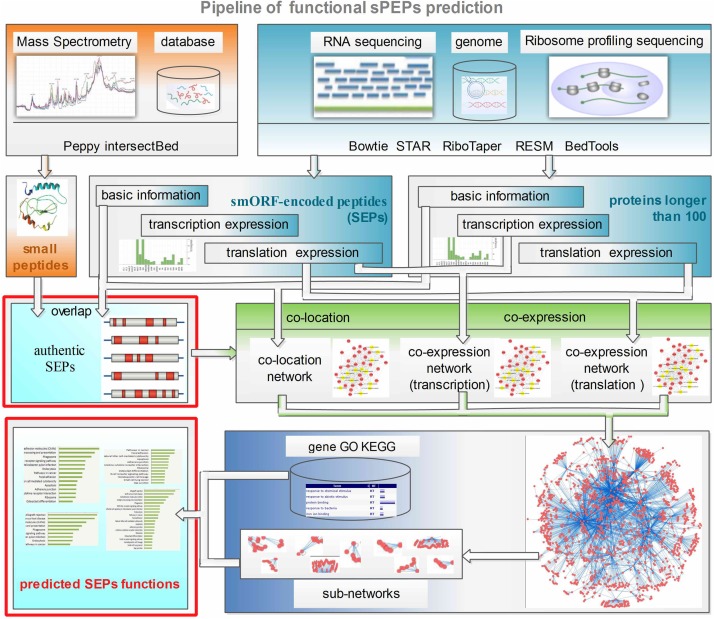
FIGURE 1.
Pipeline of FSPP. Ribo-seq, RNA-seq and mass spectrometry data are input into FSPP. This tool integrates RiboTaper, Peppy etc. to analyze the three kinds of input and gets the fundamental and expression information of translated products. FSPP uses the overlap results of MS and Ribo-seq as the authentic target SEPs. Co-expression relevance and co-location information are used to build up the relation networks. FSPP selects the sub-networks which are significantly related with the target SEPs and annotate the SEPs’ functions with the help of the known neighbors. FSPP, functional smORF-encoded peptides predictor; MS, mass spectrometry; SEPs, smORF-encoded peptides.