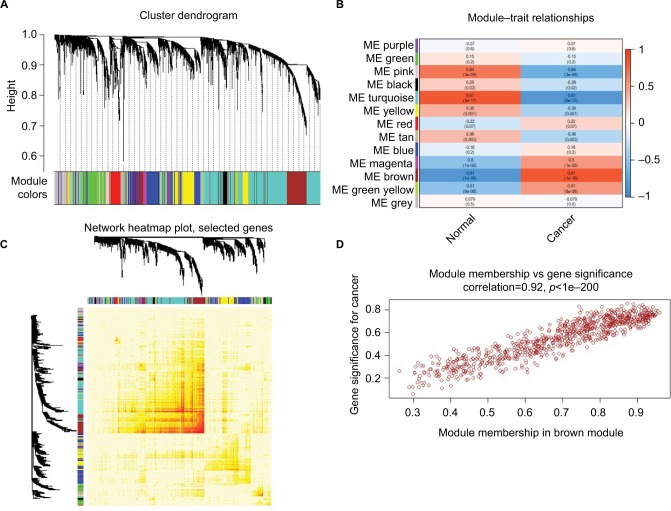
Figure 3.
WGCNA of GSE29570 and GSE89657.
Notes: (A) Six-thousand-sixty genes were assigned to one of 13 modules, including Module gray, with a cutoff of powers =6. The top image shows a gene dendrogram, and the bottom image shows the gene modules with different colors. (B) Correlation between modules and traits. The upper number in each cell refers to the correlation coefficient of each module in the trait, and the lower number is the corresponding p-value. Among them, the Brown and Turquoise modules were the most relevant modules with cancer traits. (C) A heatmap of 1,000 genes was selected at random. The intensity of the red color indicates the strength of the correlation between pairs of modules on a linear scale. (D) A scatter plot of GS for cervical cancer versus the MM in the Brown module. Intramodular analysis of the genes found in the Brown module, which contains genes that have a high correlation with cervical cancer, with p<1e-200 and correlation =0.92.
Abbreviations: GS, gene significance; MM, module membership; WGCNA, Weighted gene coexpression network analysis.