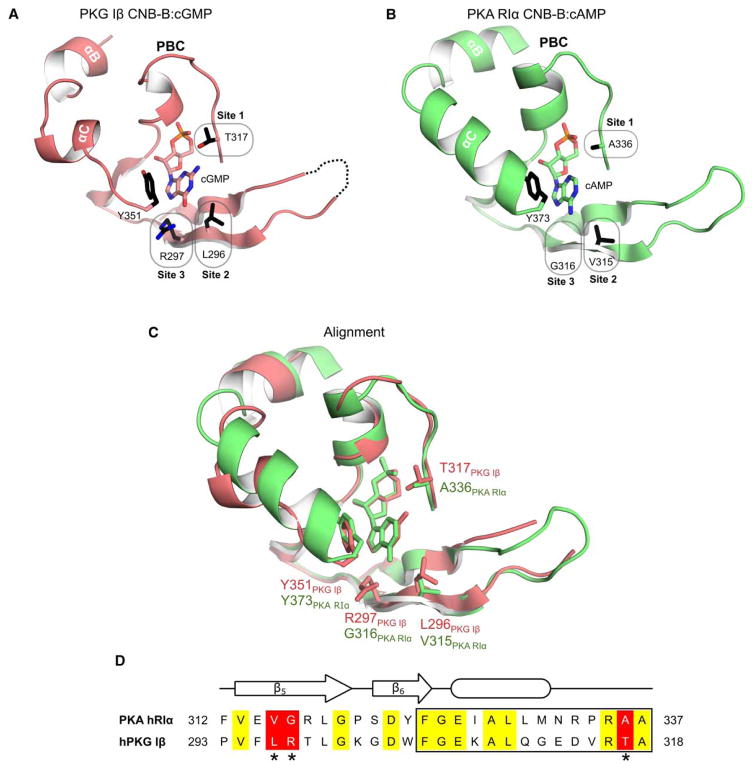
Figure 1. Structural alignment of CNB domains from PKG and PKA.
(A) Co-crystal structure of the C-terminal CNB domain (CNB-B) of PKG Iβ with cGMP (PDB code 4KU7) [20]. Key cGMP contact residues are highlighted (sites 1–3). Y351 at the C-terminal loop provides a capping interaction for cGMP. (B) Crystal structure of human PKA RIα CNB-B. Corresponding residues of sites 1–3 differ from PKG Iβ. Tyrosine 373 (Y373) is homologous to Y351 of PKG Iβ and caps the bound cAMP. (C) Structural alignment of the CNB-Bs of PKG Iβ and PKA RIα. (D) Sequence alignment from the β5 strands to the end of the PBC (box). Sequences were aligned using Clustal Omega [46,47]. Identical residues are shown in yellow. The three differing sites are shaded in red and marked with asterisks. All structure figures were generated using PyMOL (DeLano Scientific, Palo Alto, CA, U.S.A.).