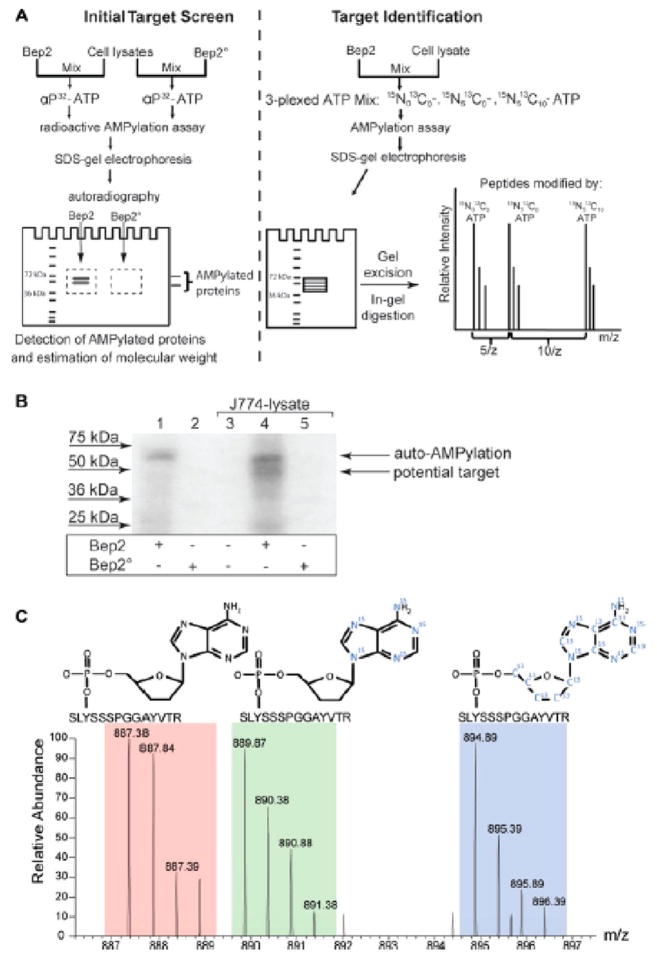
Figure 5.
Experimental strategy for the identification of in vitro AMPylated peptides using a mixture of unlabeled ATP, [15N]ATP, and [15N,13C]ATP. (A) Workflow for experimental analysis of AMPylated peptides. Briefly, AMPylation assays are performed with [32P]-labeled ATP and analyzed by PAGE to determine the molecular weight of the target proteins. Additionally, AMPylation assays are performed with a mixture of unlabeled ATP, [15N]ATP, and [15N,13C]ATP and also separated by PAGE for analysis by LC-MS/MS. [32P] radiolabeled gel is used to identify appropriate bands for excision and analysis on the [15N]ATP/[15N,13C]ATP-labeled samples. (B) Example of [32P]-labeled AMPylated protein bands identified by autoradiography. (C) Example of a MS/MS spectrum of a peptide that has been in vitro AMPylated with a mixture of unlabeled ATP, [15N]ATP, and [15N,13C]ATP. Peaks corresponding to all three forms to ATP labeling were identified. Reproduced from ref 17. Copyright 2014, with permission from John Wiley and Sons.