Figure 2. State 2 corresponds to an asymmetric trimer, in which a single CD4 molecule engages HIV-1 Env.
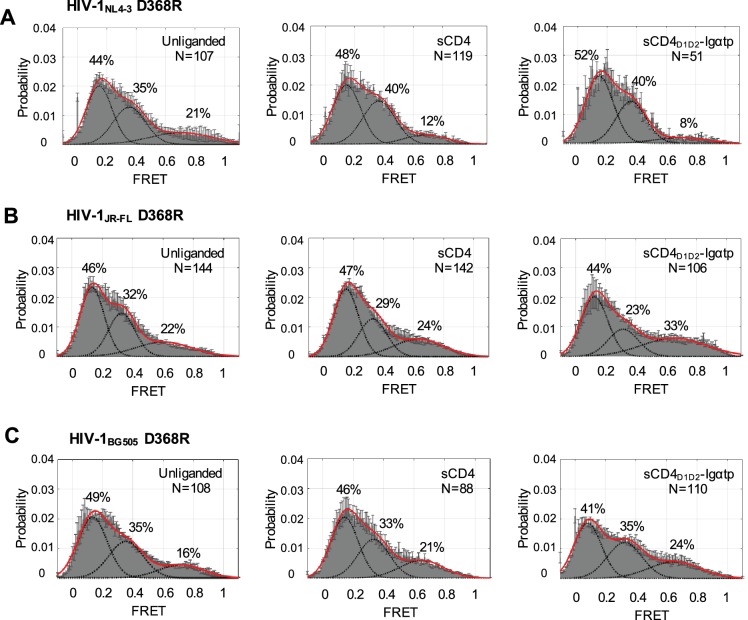
(A) Neutralization curves of WT and D368R HIVNL4-3 viruses by sCD4 and sCD4D1D2-Igαtp. Data represent three independent experiments ± standard deviation. (B) Scheme to illustrate generation of mixed HIV-1 Env trimer 1, in which the two unlabeled protomers contained the D368R mutation to prevent CD4 binding, and the CD4-binding competent WT protomer carried the donor and acceptor fluorophores (green, red stars in scheme above). (C) Scheme to illustrate generation of mixed HIV-1 trimer 2, in which sCD4 can only engage gp120 domains adjacent to the labeled domain. Given the co-transfection protocol of indicated HIV-1 plasmids, only 50% of all trimers are expected to exhibit this configuration. 25% of trimers are expected to carry D368R mutation in all three protomers and the remaining 25% would carry two CD4-binding competent protomers next to the labeled mutant gp120. (D) FRET histogram as in Figure 1 for the mixed HIV-1NL4-3 Env trimer 1. (E) FRET histogram for the mixed HIV-1NL4-3 Env trimer 2. Neutralization curves for HIV-1JR-FL (F) or HIV-1BG505 (I) were shown as in HIV-1NL4-3. FRET histograms for mixed HIV-1JR-FL (G–H) and HIV-1BG505 (J–K) Env trimer 1 and 2 are shown as HIV-1NL4-3. Viruses were incubated with sCD4 (0.1 mg/ml) or sCD4D1D2-Igαtp (0.01 mg/ml) for 30 min prior to imaging as indicated. (L) Schematic illustration of the asymmetric opening of the Env trimer. The CD4-bound conformation is in pink, and the conformational intermediate in the asymmetric trimer is in purple. The purple x indicates the D368R mutation. Green and red stars represent donor and acceptor fluorophores, respectively. Sizes of the stars represent relative change of fluorescence between donor and acceptor dyes and dotted line indicated changes of inter-dye distances.
Figure 2—figure supplement 1. D368R carrying Envs are expressed and incorporated into virions similar to wild-type.
Figure 2—figure supplement 2. Infectivity of HIV-1D368R viruses.