Fig 4. Dot1 associates with TERRA and anti-TERRA disrupts this interaction.
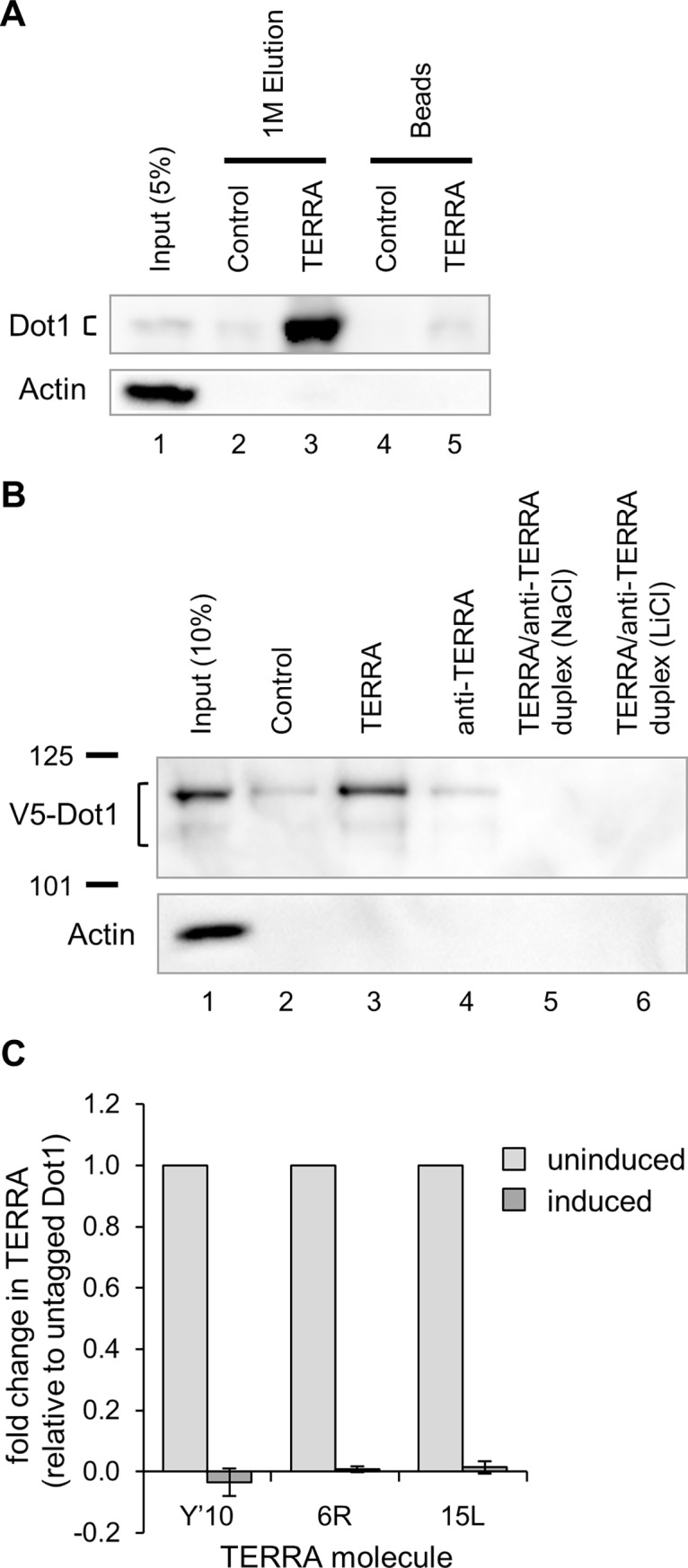
(A) TERRA-like RNA oligonucleotides but not anti-TERRA molecules can pull down native Dot1 from yeast whole cell extracts. Yeast whole cell extracts (WCE) were subject to RNA affinity purification using biotinylated TERRA or control (random sequence) RNA oligonucleotides. Bound materials were eluted with 1M NaCl in lanes 2 and 3 and then the beads were boiled in lanes 4 and 5. Lane 1 is 5% of WCE as input. Proteins were visualized by western blot with anti-Dot1 antibody and anti- -actin antibody as a control. Dot1 protein migrates near 65 kD as a doublet. (B) V5-tagged Dot1 binds TERRA and anti-TERRA prevents this interaction in vitro. Nuclear extracts were subject to RNA pulldown using the indicated RNA templates. Bound materials were eluted with 2X Laemmli buffer by boiling and assayed by western blot with antibodies specific to V5 or Actin. Lanes 5 and 6: TERRA oligonucleotides were annealed to anti-TERRA molecules under G-quadruplex permissive or minimizing conditions (NaCl or LiCl, respectively) and then were then transferred in the standard buffer used for RNA pulldown. The LiCl conditions rule out the possibility that folding of TERRA into G-quadruplexes, rather than forming duplexes with anti-TERRA, explains the loss of Dot1 binding. Lane 1 is 10% of input. Marker size in kD are indicated at left. Both isoforms of Dot1 can be seen around 110 kD. (C) V5-tagged Dot1 binds TERRA and anti-TERRA prevents this interaction in vivo. RNA immunoprecipitation was performed with V5-tagged Dot1 on yeast WCE. TERRA levels are quantified by qRT-PCR and displayed as fold change relative to an untagged Dot1 strain and to input (n = 2).
