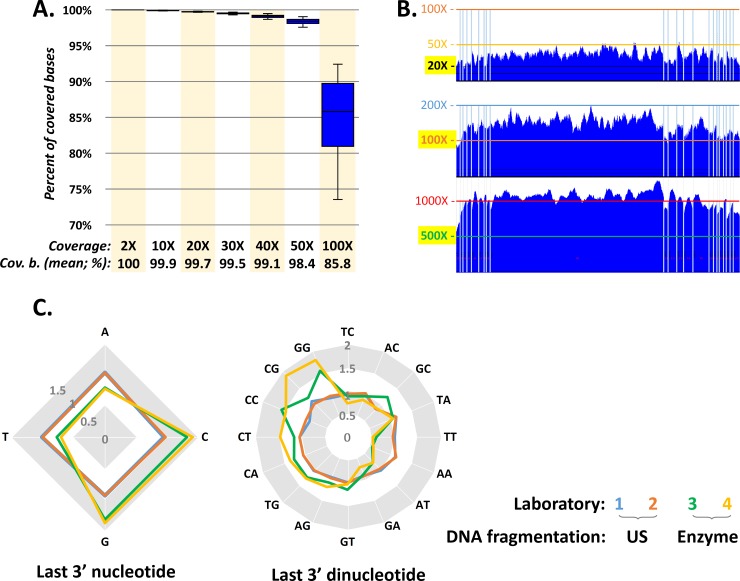
Fig 1. Coverage parameters from CZECANCA sequencing.
(A) The chart expresses the percentages of covered target bases (cov. b.) obtained from 25 analyzed samples from a standard run targeting sequencing coverage 100X. (B) The coverage (at y-axis) of BRCA1 coding sequence (NM_007294; x-axis; vertical lines represent exon boundaries) in three independent runs targeting sequencing coverages 20X, 100X, or 500X demonstrates coverage uniformity, not influenced by coverage depth. (C) The “randomness” of the DNA shearing approach using ultrasound (US) and enzymatic cleavage was compared by an analysis of the distribution of ending nucleotides and dinucleotides in reads completely mapped to the large exon 11 (chr17:41243452–41246877; 3426bp) in the BRCA1 gene, representing one of the largest continuous genomic fragments targeted by CZECANCA probes. The chart displays the relativized distribution of terminal nucleotides and dinucleotides in the analyzed region from 12 samples from each laboratory normalized to the average nucleotide and dinucleotide content of the analyzed region. The distribution of last nucleotides and dinucleotides in fragments from samples processed by US oscillate closer to a normalized value (1) than in fragments of samples prepared by the enzymatic cleavage.