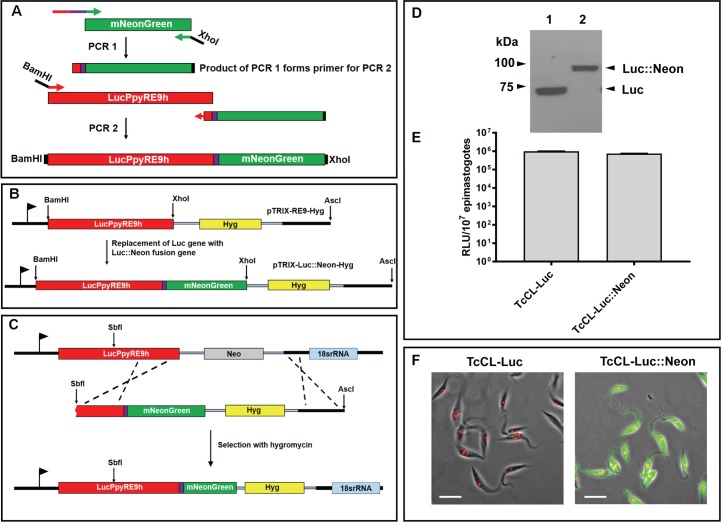
Fig 1. Generation and expression in T. cruzi of a chimeric bioluminescent–fluorescent fusion protein.
A. Amplification of the mNeonGreen ORF and flanking targeting sequences, together with the coding sequence for an 8 amino-acid spacer peptide (purple line) (Methods) (PCR1). The product was used as the downstream primer to amplify the Luc::Neon fusion fragment (PCR2). The purple box indicates the 8 amino acid spacer peptide. B. The PCR2 product was inserted into plasmid pTRIX2-RE9-Hyg [15] to create pTRIX2-Luc::Neon-Hyg. C. SbfI/AscI linearised pTRIX2-Luc::Neon-Hyg was transfected into T. cruzi CL-Luc such that integration replaces the original luciferase sequence with the fusion gene, with the G418 resistance gene (Neo) exchanged for the gene encoding hygromycin resistance (Hyg). D. Western blot probed with a polyclonal anti-luciferase antibody (Promega). Lane 1, T. cruzi CL-Luc, the original bioluminescent strain; Lane 2, T. cruzi CL-Luc::Neon parasites expressing the 88 kDa fusion protein. E. Epimastigote luciferase activity in parasite lysates (Methods). Extracts were assayed in quadruplicate, data show mean activity, and error bars indicate standard deviation. F. Epimastigotes fixed in 2% paraformaldehyde and imaged on a Zeiss LSM 510 confocal microscope. The bar indicates 10μm. Only cells carrying the fusion protein exhibit green fluorescence. DNA is stained with DAPI (coloured red on images).