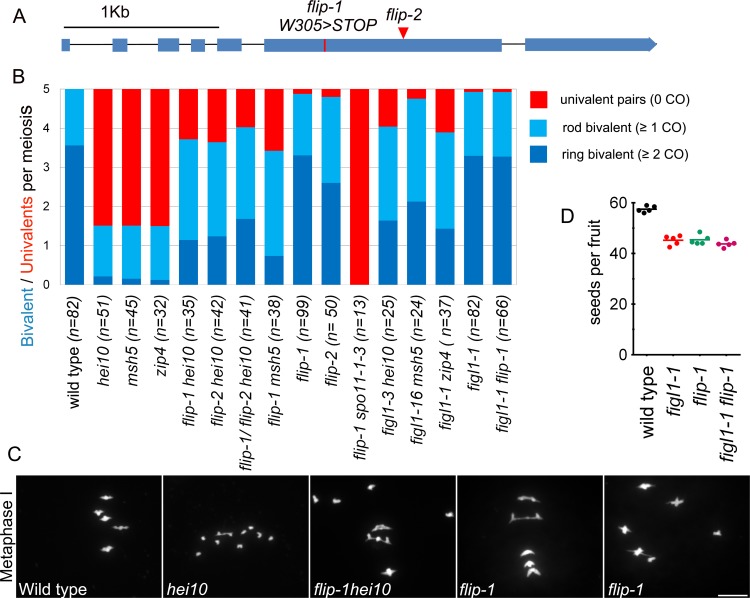
Fig 4.
Mutation in FLIP restores crossover formation in zmm mutants: A. Schematic representation of the FLIP gene (Fidgetin-Like-1 Interacting Protein). Exons appear as blue boxes. The red line and red triangle indicate the missense mutation in flip-1 and the flip-2 T-DNA insertion, respectively. B. Average number of bivalents (blue) and pairs of univalents (red) per male meiocyte at metaphase I (Fig 4C). Light blue represents rod shaped bivalents indicating that one chromosome arm has at least one CO, and one arm has no CO. Dark blue represents ring shaped bivalent indicating the presence of at least one CO on both chromosome arms. The number of cells analyzed for each genotype is indicated in brackets. C. DAPI staining of Chromosome spreads of male meiocytes at metaphase I. Scale bars 10μm. D. Fertility measured as number of seeds per fruit. Each dot represents a plant; at least 10 fruits per plant were analyzed.