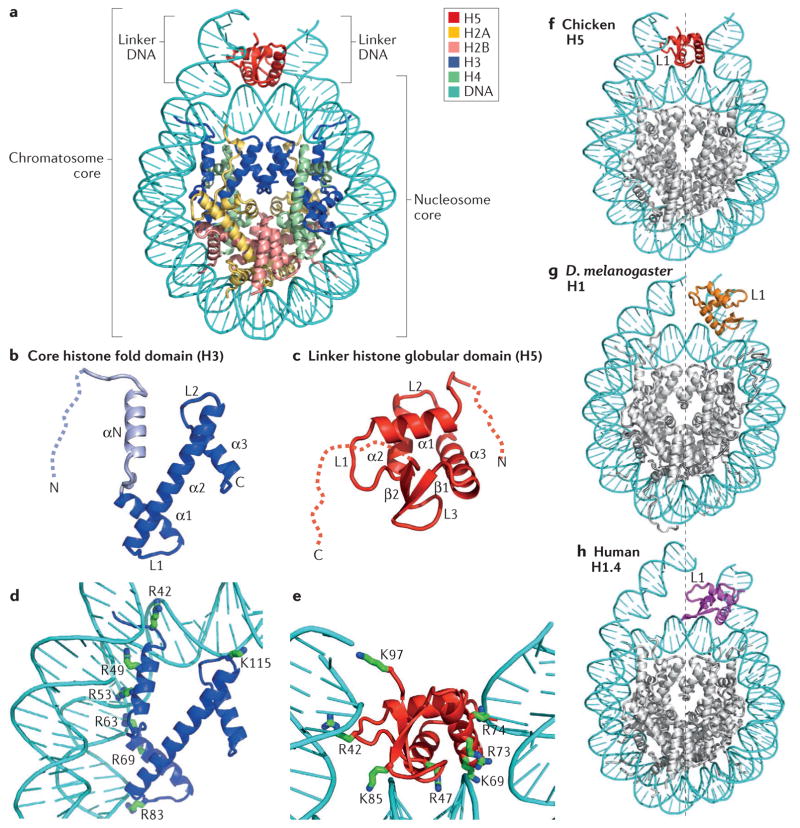
Figure 2. Structural illustration of the folded core regions of a chromatosome and representative interactions between histones and DNA.
a | The crystal structure of the chromatosome core containing the globular domain of chicken H5 (H1.0; shown in red) and fold regions of core histones (H2A, H2B, H3 and H4; all colour-coded) (Protein Data Bank identifier (PDB ID): 4QLC). The globular domain sits on the dyad of the nucleosome and interacts with both linker DNAs. b | The H3 structure from part a. The structural region from α1 to α3 (in blue) is termed the histone fold, which is shared by all core histones. The dashed line represents the intrinsically disordered histone tail. c | The structure of the folded globular domain of H5 from part a. The dashed line is used to illustrate the intrinsically disordered tails. In parts b and c, N and C indicate amino termini and carboxy termini, respectively. L indicates loop regions. d | Main interactions between DNA and the core histone H3 in the nucleosome (PDB ID: 4QLC). e | Main interactions between DNA and the globular domain of H5 (PDB ID: 4QLC). Lys (K) and Arg (R) residues that presumably form electrostatic interactions with the DNA phosphates are shown in sticks and are labelled with their residue numbers. f | The on-dyad binding mode observed in the crystal structure of the mono-nucleosome bound to the globular domain of H5 (H1.0), as in part a. g | The off-dyad binding mode observed in the NMR structural model of the mono-nucleosome bound to the Drosophila melanogaster linker histone H1 (REF. 46). h | The off-dyad binding mode observed in the cryo-electron microscopy structure of the nucleosome array containing human linker histone variant H1.4 (REF. 47). The L1 loop in the globular domain is labelled to highlight the difference in the orientation of the globular domain for each binding mode. The dashed line in parts f–h indicates the nucleosome dyad.