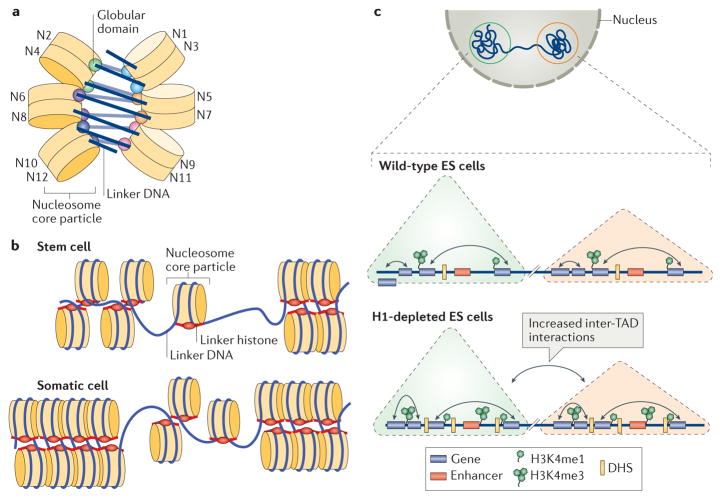
Figure 3. Roles of linker histones in chromatin folding.
a | A cartoon illustration of the cryo-electron microscopy structure of the nucleosome array condensed by human linker histone H1.4 (REF. 47). The nucleosome array structure is a twisted double helix with tetra-nucleosomes as the structural unit. The globular domain of the linker histone in each nucleosome is mainly associated with one linker DNA. The globular domains of the linker histones in the nucleosomes that interact between neighbouring tetra-nucleosome units form a dimer. The linker DNA connecting the two nucleosome core particles between neighbouring tetra-nucleosome units is not associated with the globular domain of the linker histones. The tails of the linker histones in the structure are not observed. b | Nucleosomes in the presence of linker histones are likely to form chromatosomes, which are arranged in heterogeneous groups, termed ‘nucleosome clutches’, along the chromatin fibre in a cell type-specific manner: in stem cells, smaller clutches are typically observed compared with differentiated cells. This different organization of chromatosomes corresponds to differences in chromatin compaction and heterochromatization: larger clutches are associated with heterochromatin formation. c | In interphase, the chromosome is structurally organized into distinct topologically associating domains (TADs; triangles). In H1-depleted mouse embryonic stem (ES) cells, the overall genome organization into TADs is not majorly affected. In addition, interactions (black double-headed arrows) within TADs do not change. However, within gene-dense TADs, long-range inter-TAD interactions increase. In addition, new DNase hypersensitive sites (DHSs) and sites of histone H3 Lys4 mono-methylation and trimethylation (H3K4me1 and H3K4me3, respectively) are established, indicating changes in the epigenetic landscape of the cell. Part c is adapted with permission from REF. 150, Macmillan Publishers Limited.