Figure 4. Biological functions of linker histones.
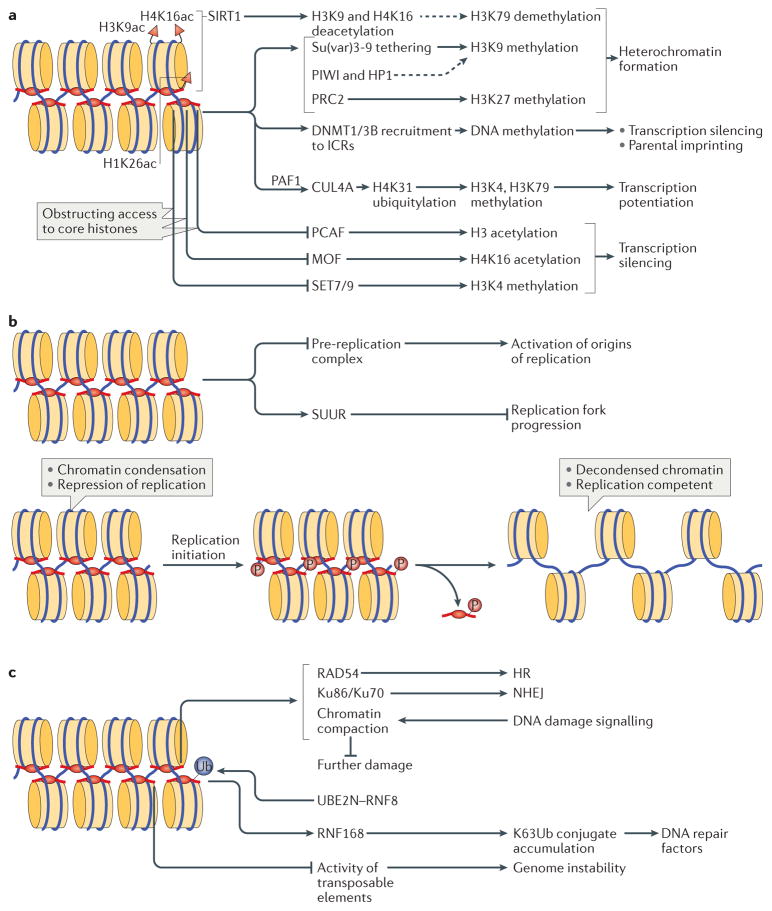
a | Linker histones (H1) are implicated in the regulation of the epigenetic landscape of the cell by interacting with several epigenetic modifiers and by regulating their recruitment to chromatin and/or activity, which affects chromatin organization. H1 acetylated at K26 (H1K26ac) is a binding partner for the deacetylase sirtuin 1 (SIRT1), which deacetylates both core histones (H3K9ac and H4K16ac) and H1; an undefined SIRT1–H1-dependent mechanism has been further linked to hypomethylation of H3 at Lys79 (H3K79). H1 is known to recruit the histone methyltransferase Su(var)3-9, as well as DNA methyltransferases DNMT1 and DNMT3B to chromatin. Furthermore, H1 interacts with PIWI proteins and heterochromatin protein 1 (HP1), modulating histone methylation and heterochromatin formation. H1 is also a substrate for methyltransferase Polycomb repressive complex 2 (PRC2) and promotes its activity. During transcription, H1 was also shown to recruit the E3 ubiquitin ligase cullin 4A (CUL4A) and RNA polymerase II-associated factor 1 (PAF1), which is necessary for CUL4A activity. By bringing CUL4A and PAF1 together, H1 promotes CUL4A-mediated ubiquitylation of H4, which further drives methylation of core histones. H1 also repels and/or interferes with the activity of several core histone modifying enzymes, including p300/CBP-associated factor (PCAF), MOF and SET7/9. b | H1-mediated mechanisms of DNA replication control. H1 represses DNA replication at the stage of replication initiation by inhibiting the assembly of the pre-replication complex and at the stage of replication fork progression by tethering the SNF2-like ATPase protein suppressor of underreplication (SUUR). In addition, H1 undergoes S phase-dependent phosphorylation (P), which results in H1 dissociation from chromatin, leading to large-scale chromatin decondensation and the activation of origins of replication. c | The roles of H1 in DNA repair, genomic stability and DNA damage signalling. H1 is involved in DNA repair via both homologous recombination (HR) and non-homologous end joining (NHEJ) through interactions with RAD54 and with Ku86 and Ku70, respectively. H1 also facilitates ubiquitin-dependent signalling at DNA double-strand breaks: H1 is ubiquitylated by the E2 ubiquitin-conjugating enzyme UBE2N and the E3 ubiquitin ligase RNF8, and recruits the E3 ubiquitin ligase RNF168 to promote accumulation of K63-linked ubiquitin conjugates, resulting in the binding of repair factors. Chromatin compaction promoted by H1 may help to limit further DNA damage. Suppression of transposable element activity by H1 also contributes to genome stability. ICRs, imprinting control regions.