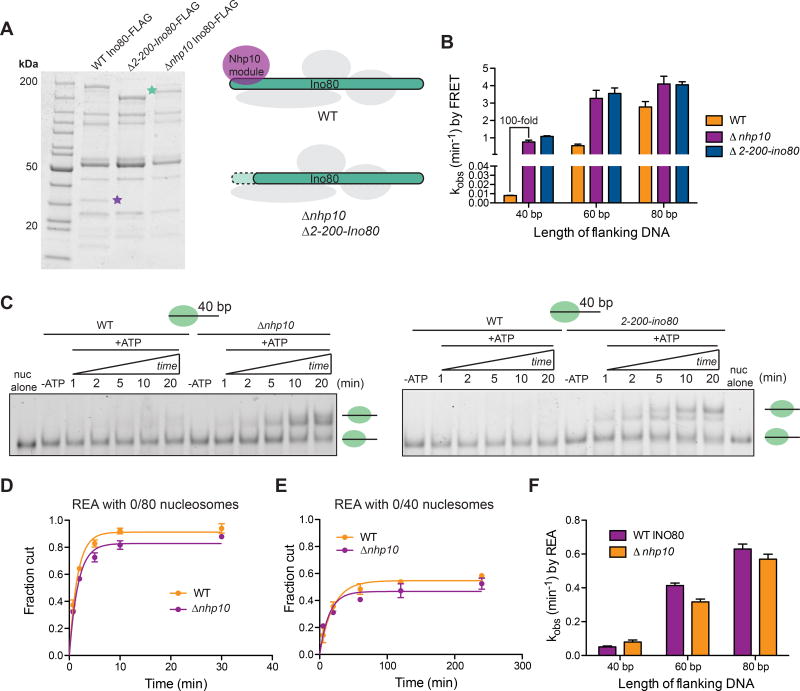
Figure 4. Deletion of nhp10 results in impaired DNA length sensing by INO80.
(A) Left, denaturing gels showing purifications of Ino80-FLAG(WT), Ino80-FLAG(Δ2-200-ino80) and Ino80-FLAG(Δnhp10). Green and purple stars represent where the Ino80 ATPase and Ies3, a major component of the Nhp10 module run on the gel, respectively. Right, schematic illustrating the compositions of mutant INO80 complexes purified from Δnhp10 and Δ2-200-ino80 strains.
(B) Remodeling rate constants measured by ensemble FRET for 0/40, 0/60 and 0/80 nucleosomes and the various INO80 mutants described in A. These assays were performed under single turnover conditions and with saturating enzyme and ATP.
(C) Native gel remodeling of 0/40 nucleosomes by INO80(Δnhp10) and INO80(Δ2-200-ino80).
These assays were performed under single turnover conditions and with saturating enzyme and ATP. Gel remodeling of 0/60 and 0/80 nucleosomes are shown in Figure S5.
(D) Quantification of fraction cut by REA on 0/80 nucleosomes, with either INO80(WT) or INO80(Δnhp10).
(E) Same as D but with 0/40 nucleosomes. For 0/60 nucleosomes, see Figure S5. Note difference in x-axis from D.
(F) Rate constants measured by REA for 0/40, 0/60 and 0/80 nucleosomes with INO80(WT) or INO80(Δnhp10). These assays were performed under single turnover conditions and with saturating enzyme and ATP. Data in (B) and (F) represent means±S.E.M. for three replicates.